Volume 11, Number 11—November 2005
Research
Norovirus Outbreaks from Drinking Water
Abstract
As part of an intensified monitoring program for foodborne disease outbreaks in Finland, waterborne outbreaks were investigated for viruses. The diagnostic procedure included analysis of patients' stool samples by electron microscopy and reverse transcription–polymerase chain reaction (RT-PCR) for noroviruses and astroviruses. When these test results were positive for a virus, the water sample was analyzed. Virus concentration was based on positively charged filters from 1-L samples. Of the total 41 waterborne outbreaks reported during the observation period (1998–2003), samples from 28 outbreaks were available for analysis. As judged by RT-PCR results from patient samples, noroviruses caused 18 outbreaks. In 10 outbreaks, the water sample also yielded a norovirus. In all but 1 instance, the amplicon sequence was identical to that recovered from the patients. The ubiquity of waterborne norovirus outbreaks calls for measures to monitor water for viruses.
Water can be a source of disease outbreaks (1). Contamination takes place almost exclusively by sewage that contains enteric pathogens, and enteric viruses that affect humans are mostly species-specific; their abundance may be explained by high concentrations in the stool of patients. Noroviruses (previously called Norwalk-like viruses) cause gastroenteritis in all age groups. Since noroviruses, unlike enteroviruses, do not easily grow in cell culture, their role became evident only in the 1990s, when specific diagnostic methods became available. Only in recent years has the vast genomic variety of noroviruses become apparent (2). A recent report (3) lists 5 genogroups and 22 genetic clusters that include mostly human but also porcine and murine viruses.
In addition to numerous community-based outbreaks, in which transmission is thought to take place from person to person, outbreaks caused by contaminated food have been frequent (4). The dominant role of noroviruses in foodborne and waterborne outbreaks has been estimated by Mead et al. (5). Several waterborne outbreaks have been detected on the basis of epidemiologic evidence (6,7), and only in 1997 did the first report of noroviruses in well water appear (8). The genome-based diagnostic procedure, i.e., reverse transcriptase–polymerase chain reaction (RT-PCR), offers a sensitive and specific tool to identify these viruses. Sequence-based identification is effective for source-tracking outbreaks, especially those caused by noroviruses, which show a highly variable nucleotide sequence even within the short amplicon produced in the polymerase region of the virus (9).
Waterborne viral outbreaks are often difficult to recognize. Illness caused by norovirus is common, and if the contamination level is low, the number of cases remains low. A rather extensive outbreak is usually required for medical personnel and authorities to recognize water as a possible source of infection (10). This report includes virologic analyses of Finnish waterborne outbreaks during a 6-year period. We describe an improved procedure to identify water as the source of viral outbreaks.
Reporting of foodborne and waterborne outbreaks in Finland was reorganized and intensified in 1997; new regulations emphasized that all suspected cases should be immediately reported to the National Public Health Institute (KTL). Recommendations were given for properly collecting both patient and environmental samples. The functions of local outbreak investigation teams were clarified and included training in conducting epidemiologic surveys. Laboratory performance was improved by including options for viral and protozoan diagnostics from both patient and environmental samples. All cases in which water was suspected as the source of the outbreak were reported to KTL. Sampling recommendations included 3–10 representative patient stool samples. Water samples, raw water, and when appropriate, tap water from different parts of the distribution network were collected immediately. Despite recommendations, not all outbreaks were investigated for viruses. The criteria for establishing an outbreak as waterborne were according to the English classification (grades A–D) (11).
In total, 271 patient samples from 25 outbreaks were analyzed for viruses. The range of fecal samples obtained from each outbreak was 2–69 (mean 11). A 10% fecal suspension in 0.05 mol/L Tris-HCl, 0.1 mol/L NaCl, 1 mmol/L CaCl2, pH 7.4, was used for RNA extraction.
A total of 73 water samples from 27 outbreaks were analyzed; 1- to 2-L water samples, collected in clean glass or plastic bottles, were concentrated as described by Gilgen et al. (12). The 1-L samples were run through a positively charged disk membrane filter (diameter 47 mm, pore size 45 μm; AMF-Cuno, Zetapor, Meriden, CO, USA) with or without a fiberglass prefilter. After the elution step in 50 mmol/L glycine buffer, pH 9.5, containing 1% beef extract, the eluate was rapidly neutralized with HCl. The volume was further reduced to ≈100 μL with a microconcentrator (Centricon-100, Amicon, Beverly, MA, USA). This sample was used for RNA extraction and PCR as described (10).
RNA extraction and RT-PCR for the norovirus polymerase region were performed as described (13). Briefly, RNA was extracted by using phenol- and guanidine thiocyanate–containing Tripure reagent (Roche, Indianapolis, IN, USA) and precipitated with ethanol. Viral RNA was transcribed to cDNA, and DNA amplification was performed in separate tubes for norovirus genogroups I and II (GI and GII) by manual PCR with primers Nvp110 (14) and N69 (15), and Nvp110 and NI (16), respectively. From 2002 on, the forward primers for the genogroups were modified as KA1 (5´-GANGGCCTSCCMTCWGGNTT-3´) and KA2 (5´-TGGAATTCNATHGCCCAYTGG-3´). The amplicons were visualized by electrophoresis in an agarose gel, hybridized by a probe panel, and used for nucleotide sequencing.
Sequencing was performed manually (Sequenase, version 2.0 DNA sequencing kit, USB, Cleveland, OH, USA) as described (13). Sequence analysis was performed by programs SeqApp and ClustalW. Our sequences were aligned with the following EMBL/GenBank noroviruses: Southampton/91/UK (L07418), Norwalk/68/US (M87661), Malta (AJ277616), Melksham/94/UK (X81879), Hawaii/76/US (U07611), Lordsdale/93/UK (X86557), GIIb (AY7732101), GIId (AF312728), and murine norovirus (AY228235). For nucleotide sequences for Hillingdon/94/UK and Grimsby/95/UK, see Vinje et al (17); sequence of Lord Harris comes from the sequence database of the European network (9,18). GenBank accession numbers for nucleotide sequences of this study are AY958213–9 for GI and AY958204–12 for GII noroviruses.
Description of Outbreaks and Viral Findings
In total, 41 waterborne outbreaks (3–11 per year) were registered in Finland from 1998 to 2003. Of these, 28 (61%) were investigated for viruses. In 24 outbreaks both water and patient samples were available for analysis; in 3 outbreaks only water was available, and in 1 outbreak only patient samples were available for analysis. Samples for viral analysis were not obtained from the remaining 13 outbreaks. Analysis was performed by RT-PCR. Patient samples were also screened by electron microscopy for other enteric viruses and analyzed by RT-PCR for astroviruses. For water samples, a concentration method according to Gilgen et al. (12) was established, starting from the volume of 1 L. In most cases, water samples were analyzed only for noroviruses. The most prominent viruses that caused waterborne outbreaks were noroviruses (18 outbreaks). Rotavirus caused 1 waterborne outbreak, and no viruses were found in 9 epidemics. Bacterial findings will be published elsewhere.
The 18 waterborne norovirus outbreaks are summarized in Table 1. In every year except 2001, several norovirus outbreaks occurred in Finland. During the study period, 6 large norovirus epidemics with >200 cases were encountered. In the largest epidemics, >10,000 persons were exposed, and 2,000–5,500 cases occurred; in addition, 7 medium-sized (40–100 cases) and 5 small outbreaks (<20 cases) were caused by noroviruses.
Most norovirus contaminations occurred in groundwater systems, which are used most commonly in Finland. In 3 instances, surface, lake, or river water was used. Of the ground water epidemics, 8 occurred in public communal systems and 7 in private ground water wells. Typically rental cottages or different kinds of camping grounds with their own wells were affected.
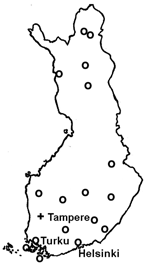
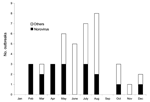
The geographic distribution of the waterborne norovirus outbreaks is shown in Figure 1. Outbreaks occurred all over the country, from the southern archipelago to the northernmost parts of Finland. Seasonal risk for waterborne norovirus outbreak seemed to be approximately equal (Figure 2). Half (20 of 41) of the waterborne epidemics occurred in summer, and norovirus outbreaks (11 of 15) were most common in late winter to spring (February–May). In fact most outbreaks in winter were caused by noroviruses, while in summer they were mainly caused by bacteria.
Detailed Analysis of Noroviruses
Noroviruses from 16 outbreaks (E1–E16) were further characterized by sequence analysis of amplicons, from which the genotype was also deduced (Table 2). Noroviruses appeared in the patient samples in all 16 outbreaks and in water samples of 10 epidemics. Coliforms were also present in 9 epidemics, whereas in 7 outbreaks, no indication of microbiologic contamination was seen. Most outbreaks were caused by a single norovirus strain/genotype (11 epidemics); >1 virus was found more often in large outbreaks than in small ones. GII noroviruses were only slightly more common than GI (7 vs. 5 outbreaks). Of the 10 epidemics with positive water samples, equal numbers of GI and GII genotypes were detected.
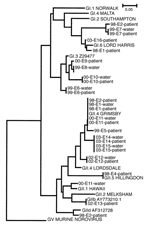
In all but 1 of these outbreaks, the same norovirus genotype found in water samples also appeared in patient samples. The only exception was epidemic E11, in which 2 norovirus sequences, GII.1 and GII.4, were detected in the water sample, but only type GII.4 was detected in the patient samples. Not only the viral genotype but also the entire amplicon sequence were identical in each outbreak (Figure 3). Two norovirus genogroup I types, GI.3 (Birmingham) and GI.6 (Sindlesham, Hesse), were found; 1 GI sequence (outbreak E3) remained undetermined.
In the GII outbreaks, at least 4 different genotypes were found in patient or water samples. The most common genotype was GII.4 (Bristol, Lordsdale), found in water samples of 4 epidemics, and beginning in 2003, it was the new variant type (20). The established genotypes GII.1 (Hawaii) and GII.5 (Hillingdon) were also detected in some outbreaks, along with some potentially new genotypes or sequences that did not cluster well in any of the established genotypes, such as GIId (Upinniemi) and GIIb. As Figure 3 shows, in most outbreaks a virus with a unique amplicon sequence was recovered, even when it belonged to the same genotype as viruses in the other waterborne outbreaks. Norovirus genotype GII.4 was the only exception, and a longer nucleotide sequence likely would have shown some genetic differences (detected between sequences of epidemics E1 and E11; data not shown).
As part of the improved and intensified outbreak surveillance system in Finland, we have identified waterborne viral outbreaks since 1998. In a relatively brief period, during which norovirus diagnostics have been available for patient as well as environmental samples, a considerable number of waterborne norovirus outbreaks have been detected.
That Finland has >1,300 water treatment plants may in part explain the numerous outbreaks. Many of these plants still use surface water (lakes or rivers) as raw water. Inadequate disinfection is then the most common reason for waterborne epidemics, as was the case in outbreak E1 (10). At risk also are water plants that use groundwater and no disinfection. In Finland, snow melts in spring while the ground is still frozen, which leads to surface runoffs and flooding. Breaks in sewer lines in the vicinity of a well caused several large waterborne outbreaks. Poor sewage disposal also caused many small waterborne outbreaks in private homes or rental cottages.
The large number of genetically distinct norovirus genotypes has been advantageous in investigating waterborne epidemics. Although the short amplicon sequence does not definitively show that 2 viruses are identical, for the purpose of source tracing it seems adequate. In this study, a unique viral sequence appeared in most norovirus outbreaks, and viruses from patients and water in a particular outbreak showed identical sequences. The success in most outbreaks in identifying a norovirus with the same sequence from patients and water may be due to the fact that the outbreaks have taken place in small communities. In large waterborne outbreaks, usually >1 norovirus strain and often other viruses and microbes are causative agents.
Both norovirus genogroups occurred in waterborne epidemics. In a 5-year study (1998–2002) in Finland, GII outbreaks clearly outnumbered those caused by GI noroviruses (86.9% vs. 13.1%) (21). In waterborne outbreaks, however, nearly half were caused by GI viruses. Some differences may occur in stability as well as ability to spread from person to person among viruses representing different genotypes. Type GI.3, the most common GI genotype in water samples, was also the most frequent GI type in community outbreaks (21). Viruses of this genotype have caused waterborne outbreaks in the United States in 2001 (22) and in the Netherlands (23).
As might be expected, keeping in mind its ubiquity (24,25), the GII.4 genotype was present in several waterborne outbreaks, and in Finland it has been the most frequent genotype in all outbreaks. The GII.4 new variant emerged in Finland in June 2002, and in the following year 2 waterborne outbreaks were caused by this new variant (20). Another emerging genotype, GIIb, found in Finland in 2001, a year later than in southern parts of Europe, was a causative agent in a waterborne outbreak in 2002. A waterborne outbreak in Sweden caused by this genotype has recently been reported (26).
Environmental virology of human pathogen detection has a rather limited history. A classic case is the monitoring of polioviruses in sewage (27). This method, based on a cell culture technique, is sensitive in detecting circulating wild poliovirus. Further efforts in environmental virology were lacking for many years, mainly because suitable methods were absent. Only after gene amplification techniques were introduced could a tool be developed to successfully detect norovirus in environmental samples (8,10). In recent years, an increasing number of reports have described waterborne norovirus outbreaks through contaminated drinking or recreational water (22,23,28,29).
National recommendations for volumes of water to be tested vary between tens and hundreds of liters. Such volumes pose a serious practical problem for the testing laboratory. For viral detection by RT-PCR, a smaller volume (1 L) is preferred, as suggested by Gilgen et al (12). Independent of the concentration method, the increase in RT-PCR inhibitors usually sets limits on the water concentration. Sensitive methods are needed to detect viruses in environmental samples. Recent reports on the applicability and sensitivity of real-time RT-PCR (30–32) for noroviruses also offer new possibilities to enhance its sensitivity. Another factor is that the test then becomes more rapid, which is essential in monitoring water quality, particularly in epidemic situations. The third advantage is that a quantitative estimate of the contamination level is obtained.
Microbial risks from water are recognized, with much emphasis on risk assessment (33). Assessment of water, however, depends on indicator organisms, such as coliforms or enterococci, whose survival in water is shorter than that of enteric viruses, especially norovirus and hepatitis A virus. Therefore, viruses can easily be harbored in "microbiologically immaculate" water (34,35). In situations in which a well is contaminated by sewage, coliforms are nearly always found. When sewage is released into lake water that serves as raw water downstream, indicator organisms may no longer be detectable, but noroviruses can still be present and cause illness. This sequence of events probably led to the first outbreak we examined (E1) (10).
When water plants use surface water, the contamination may be short-lived and may have vanished by the time the outbreak is detected. A "rolling sample" system might be used in which samples are collected in water plants at risk for contamination at regular intervals (e.g., daily, weekly) and stored at 4°C. Unless signs of an outbreak appear, the samples can be discarded at the same pace that new ones are collected. In case of contamination, water samples would be available for analyses.
The evidence presented here together with several recent reports mentioned above show the role of viruses as contaminants of drinking water. In Finland, the finding that noroviruses frequently cause waterborne outbreaks has led to authorities' increased awareness of viral risks. As a consequence, laboratory techniques have been improved, and the capacity for analyzing environmental samples, especially water, has increased. Legislative measures for viral monitoring as part of the microbial risk assessment in drinking water production should be seriously considered.
Dr Maunula is a microbiologist at the Department of Food and Environmental Hygiene of the University of Helsinki, Finland. Her main research interest is molecular epidemiology of enteric viruses, especially noroviruses and rotaviruses. In recent years she has concentrated on viruses in environmental rather than in clinical samples.
Acknowledgments
We thank all the persons involved in water analyses as well as the PCR laboratory of the Department of Virology of HUCS Laboratory Diagnostics (now HUSLAB).
This study was supported in part by research grants from the Academy of Finland (42675/98) and from a European Commission contract (Food-borne Viruses in Europe, QLK1-1999-00594).
References
- Wyn-Jones A, Sellwood J. Enteric viruses in the aquatic environment. J Appl Microbiol. 2001;91:945–62. DOIPubMedGoogle Scholar
- Green KY, Chanock M, Kapikian AZ. Human caliciviruses. In: Knipe D, editor. Fields virology. 4th ed. Philadelphia: Lippincott, Williams and Wilkins; 2001. p. 841–74.
- Vinje J, Hamidjaja RA, Sobsey MD. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J Virol Methods. 2004;116:109–17. DOIPubMedGoogle Scholar
- Koopmans M, von Bonsdorff CH, Vinje J, de Medici D, Monroe S. Foodborne viruses. FEMS Microbiol Rev. 2002;26:187–205.PubMedGoogle Scholar
- Mead P, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. DOIPubMedGoogle Scholar
- Kaplan JE, Goodman RA, Schonberger LB, Lippy EC, Gary GW. Gastroenteritis due to Norwalk virus: an outbreak associated with a municipal water system. J Infect Dis. 1982;146:190–7. DOIPubMedGoogle Scholar
- Blackburn BG, Craun GF, Yoder JS, Hill V, Calderon RL, Chen N, Surveillance for waterborne-disease outbreaks associated with drinking water—United States, 2001–2002. MMWR Surveill Summ. 2004;53:23–45.PubMedGoogle Scholar
- Beller M, Ellis A, Lee SH, Drebot MA, Jenkerson SA, Funk E, Outbreak of viral gastroenteritis due to a contaminated well. International consequences. JAMA. 1997;278:563–8. DOIPubMedGoogle Scholar
- Vinje J, Vennema H, Maunula L, von Bonsdorff CH, Hoehne M, Schreier E, International collaborative study to compare reverse transcriptase PCR assays for detection and genotyping of noroviruses. J Clin Microbiol. 2003;41:1423–33. DOIPubMedGoogle Scholar
- Kukkula M, Maunula L, Silvennoinen E, von Bonsdorff CH. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J Infect Dis. 1999;180:1771–6. DOIPubMedGoogle Scholar
- PHLS Communicable Disease Surveillance Centre. Strength of association between human illness and water: revised definitions for use in outbreak investigation. Commun Dis Rep CDR Wkly. 1996;6:65,8.
- Gilgen M, Germann D, Lüthy J, Hübner P. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int J Food Microbiol. 1997;•••:189–99. DOIPubMedGoogle Scholar
- Maunula L, Piiparinen H, von Bonsdorff CH. Confirmation of Norwalk-like virus amplicons after RT-PCR by microplate hybridization and direct sequencing. J Virol Methods. 1999;83:125–34. DOIPubMedGoogle Scholar
- Le Guyader F, Estes MK, Hardy ME, Neill FH, Green J, Brown DW, Evaluation of a degenerate primer for the PCR detection of human caliciviruses. Arch Virol. 1996;141:2225–35. DOIPubMedGoogle Scholar
- Wang J, Jiang X, Madore HP, Gray J, Desselberger U, Ando T, Sequence diversity of small, round-structured viruses in the Norwalk virus group. J Virol. 1994;68:5982–90.PubMedGoogle Scholar
- Green J, Gallimore CI, Norcott JP, Lewis D, Brown DW. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J Med Virol. 1995;47:392–8. DOIPubMedGoogle Scholar
- Vinje J, Green J, Lewis DC, Gallimore CI, Brown DW, Koopmans MP. Genetic polymorphism across regions of the three open reading frames of "Norwalk-like viruses.". Arch Virol. 2000;145:223–41. DOIPubMedGoogle Scholar
- Koopmans M, Vennema H, Heersma H, van Strien E, van Duynhoven Y, Brown D, Early identification of common-source foodborne virus outbreaks in Europe. Emerg Infect Dis. 2003;9:1136–42.PubMedGoogle Scholar
- Kuusi M, Nuorti JP, Maunula L, Miettinen I, Pesonen H, von Bonsdorff CH. Internet use and epidemiologic investigation of gastroenteritis outbreak. Emerg Infect Dis. 2004;10:447–50.PubMedGoogle Scholar
- Lopman B, Vennema H, Kohli E, Pothier P, Sanchez A, Negredo A, Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet. 2004;363:682–8. DOIPubMedGoogle Scholar
- Maunula L, von Bonsdorff C-H. Norovirus genotypes causing gastroenteritis outbreaks in Finland 1998–2002. J Clin Virol. Epub 2005 May 20.PubMedGoogle Scholar
- Parshionikar SU, Willian-True S, Fout GS, Robbins DE, Seys SA, Cassady JD, Waterborne outbreak of gastroenteritis associated with a norovirus. Appl Environ Microbiol. 2003;69:5263–8. DOIPubMedGoogle Scholar
- Hoebe CJ, Vennema H, Husman AM, van Duynhoven YT. Norovirus outbreak among primary schoolchildren who had played in a recreational water fountain. J Infect Dis. 2004;189:699–705. DOIPubMedGoogle Scholar
- Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. Identification of a distinct common strain of "Norwalk-like viruses" having a global distribution. J Infect Dis. 1999;179:1334–44. DOIPubMedGoogle Scholar
- Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Epidemiologic and molecular trends of "Norwalk-like viruses" associated with outbreaks of gastroenteritis in the United States. J Infect Dis. 2002;186:1–7. DOIPubMedGoogle Scholar
- Nygard K, Torven M, Ancker C, Knauth SB, Hedlund KO, Giesecke J, Emerging genotype (GGIIb) of norovirus in drinking water, Sweden. Emerg Infect Dis. 2003;9:1548–52.PubMedGoogle Scholar
- Lapinleimu K, Stenvik M. The efficacy of polio vaccination in Finland. Dev Biol Stand. 1978;41:137–9.PubMedGoogle Scholar
- Boccia D, Tozzi AE, Cotter B, Rizzo C, Russo T, Buttinelli G, Waterborne outbreak of Norwalk-like virus gastroenteritis at a tourist resort, Italy. Emerg Infect Dis. 2002;8:563–8.PubMedGoogle Scholar
- Anderson AD, Heryford AG, Sarisky JP, Higgins C, Monroe SS, Beard RS, A waterborne outbreak of Norwalk-like virus among snowmobilers—Wyoming, 2001. J Infect Dis. 2003;187:303–6. DOIPubMedGoogle Scholar
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–57. DOIPubMedGoogle Scholar
- Hohne M, Schreier E. Detection and characterization of norovirus outbreaks in Germany: application of a one-tube RT-PCR using a fluorogenic real-time detection system. J Med Virol. 2004;72:312–9. DOIPubMedGoogle Scholar
- Beuret C. Simultaneous detection of enteric viruses by multiplex real-time RT-PCR. J Virol Methods. 2004;115:1–8. DOIPubMedGoogle Scholar
- World Health Organization. Guidelines for drinking-water quality. 3rd ed [monograph on the Internet]. 2004 [cited 2005 Sep 8]. Available from http://www.who.int/water_sanitation_health/dwq/gdwq3/en/
- Payment P, Franco E, Fout GS. Incidence of Norwalk virus infections during a prospective epidemiological study of drinking water-related gastrointestinal illness. Can J Microbiol. 1994;40:805–9. DOIPubMedGoogle Scholar
- Lees D. Viruses and bivalve shellfish. Int J Food Microbiol. 2000;59:81–116. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 11, Number 11—November 2005
| EID Search Options |
|---|
|
|
|
|
|
|