Volume 11, Number 6—June 2005
Research
Global Spread of Vancomycin-resistant Enterococcus faecium from Distinct Nosocomial Genetic Complex
Figure 1
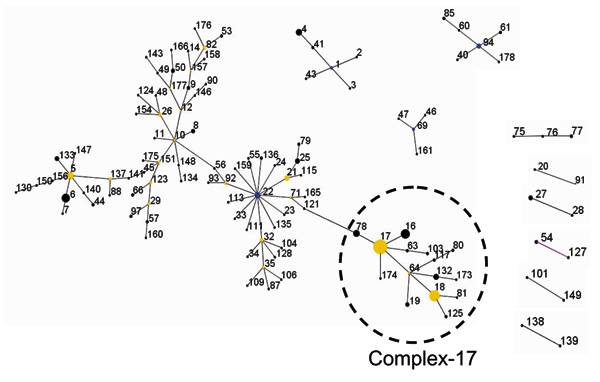
Figure 1. . Clustering of 175 sequence types representing 411 isolates with eBURST (25). This algorithm identifies the founder of a complex or genogroup of related sequence types (ST) and subsequent patterns of evolutionary descent. The primary founder, indicated in blue, of a complex is defined as the ST with the largest number of single locus variants (SLVs). Larger complexes may contain secondary founders of additional lineages that have a number of SLVs of their own. These secondary founders are indicated in yellow. Numbers correspond to ST numbers. The area of each circle corresponds to the number of isolates of the ST. All complexes (major and minor) are shown. In addition, 57 STs did not group into any of the complexes and were considered singletons (STs 13, 15, 30, 31, 36, 37, 38, 39, 42, 51, 52, 58, 59, 62, 65, 67, 68, 70, 72, 73, 74, 83, 84, 86, 89, 95, 96, 98, 99, 100, 102, 105, 107, 108, 110, 112, 114, 116, 118, 126, 129, 131, 142, 144, 145, 152, 153, 155, 162, 163, 164, 167, 168, 169, 170, 171, 172). The "epidemic" genetic complex-17 derived from secondary founder ST-17 is indicated. A measurement of statistical confidence in each of the assigned primary founders is made by a bootstrap resampling procedure (25).The predicted primary founders of the complexes 22, 94, 1, and 69 have a bootstrap value of 73%, 84%, 85%, and 59%, respectively.