Volume 11, Number 7—July 2005
Research
Primate-to-Human Retroviral Transmission in Asia
Figure 3
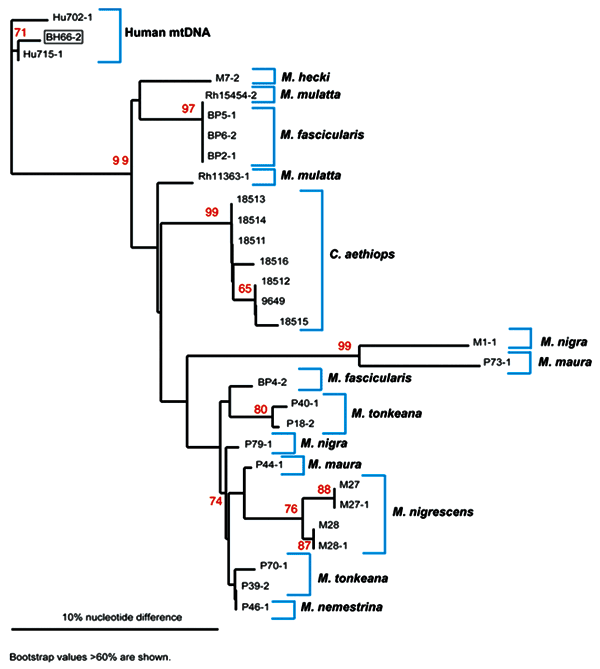
Figure 3. . Phylogenetic analysis of mitochrondrial (mt) DNA from nonhuman primates and humans. mtDNA was amplified and sequenced from the simian foamy virus–infected person (BH66), 2 human controls (Hu702 and Hu715), M. mulatta (Rh15454, 18511,18512, 18513,18514,18515, 11363, 9649), M. fascicularis (BP2, 4, 5, 6), M. nemestrina (P46), M. tonkeana (P18,39,40), M. maura (P44, 73), M. nigra (P79, M1); M. nigrescens (M27, 28), and M. hecki (M7). The mtDNA tree was created with the neighbor-joining method with the Phylip program (DNAdist; Neighbor). Bootstrap replicates were 1,000. Bootstrap values were calculated by using Seqboot, DNAdist, Neighbor, and Consense (PHYLIP programs). Bootstrap values >60% are shown. The mtDNA tree was plotted in Treeview. This analysis suggests that BH66 was of human origin. Although the phylogenetic tree constructed with mtDNA from a variety of monkey samples can be used to distinguish human from monkey mtDNA, a large number of nuclear mtDNA sequences, have evolved as pseudogenes (36). These sequences can be highly divergent from mtDNA and resulted in some ambiguity as mtDNA amplified from several monkeys did not group with other members of the same species. Because of the nature and variability of these sequences, definitive conclusions about mtDNA phylogenies could not be determined; however, mtDNA trees were still useful for determining the origin of mtDNA material.
References
- Wolfe ND, Escalante AA, Karesh WB, Kilbourne A, Spielman A, Lal AA. Wild primate populations in emerging infectious disease research: The missing link? Emerg Infect Dis. 1998;4:149–58. DOIPubMedGoogle Scholar
- Weiss RA. Cross-species infections. Curr Top Microbiol Immunol. 2003;278:47–71. DOIPubMedGoogle Scholar
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–41. DOIPubMedGoogle Scholar
- Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, Hahn BH. The origins of acquired immune deficiency viruses: where and when? Philos Trans R Soc Lond B Biol Sci. 2001;356:867–76. DOIPubMedGoogle Scholar
- Murphy FA. A perspective on emerging zoonoses. In: Burroughs T, Knobler S, Leberberg J, editors. The emergence of zoonotic diseases: Understanding the impact on animal and human health. Washington: National Academy Press; 2002. p. 1–10.
- Khabbaz RF, Heneine W, George JR, Parekh B, Rowe T, Woods T, Brief report: infection of a laboratory worker with simian immunodeficiency virus. N Engl J Med. 1994;330:172–7. DOIPubMedGoogle Scholar
- Gessain A, Mahieux R. Epidemiology, origin and genetic diversity of HTLV-1 retrovirus and STLV-1 simian affiliated retrovirus. Bull Soc Pathol Exot. 2000;93:163–71.PubMedGoogle Scholar
- Vandamme AM, Salemi M, Desmyter J. The simian origins of the pathogenic human T-cell lymphotropic virus type I. Trends Microbiol. 1998;6:477–83. DOIPubMedGoogle Scholar
- Gessain A, Vernant JC, Barin F, Maurs L, Gout O, Calendar A, Antibodies to human T-lymphotropic virus type-1 in patients with tropical spastic paraparesis. Lancet. 1985;2:407–10. DOIPubMedGoogle Scholar
- Lerche NW, Switzer WM, Yee JL, Shanmugam V, Rosenthal AN, Chapman LE, Evidence of infection with simian type D retrovirus in persons occupationally exposed to nonhuman primates. J Virol. 2001;75:1783–9. DOIPubMedGoogle Scholar
- Switzer WM, Bhullar V, Shanmugam V, Cong M, Parekh B, Lerche NW, Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J Virol. 2004;78:2780–9. DOIPubMedGoogle Scholar
- Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Naturally acquired simian retrovirus infections among Central African hunters. Lancet. 2004;363:932–7. DOIPubMedGoogle Scholar
- Brooks JI, Erling R, Pilon RG, Smith JW, Switzer WM, Sandstrom PA. Cross-species retroviral transmission from macaques to human beings. Lancet. 2002;360:387–8. DOIPubMedGoogle Scholar
- Sandstrom PA, Phan KO, Switzer WM, Fredeking T, Chapman L, Heneine W, Simian foamy virus infection among zoo keepers. Lancet. 2000;355:551–2. DOIPubMedGoogle Scholar
- Calattini S, Mauclere P, Torevoye P, Froment A, Saib A, Gessain A. Interspecies transmission of simian foamy viruses from chimpanzees and gorillas to Bantous and Pygmees hunters in Southern Cameroon. Proceedings of the 5th International Foamy Virus Conference; 2004 Jul 9–11;Wuerzburg, Germany.
- Renne R, Friedl E, Schweizer M, Fleps U, Turek R, Neumann-Haefelin D. Genomic organization and expression of simian foamy virus type 3 (SFV-3). Virology. 1992;186:597–608. DOIPubMedGoogle Scholar
- Mergia A, Shaw KE, Pratt-Lowe E, Barry PA, Luciw PA. Simian foamy virus type 1 is a retrovirus which encodes a transcriptional transactivator. J Virol. 1990;64:3598–604.PubMedGoogle Scholar
- Hussain AI, Shanmugam V, Bhullar VB, Beer BE, Vallet D, Gautier-Hion A, Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology. 2003;309:248–57. DOIPubMedGoogle Scholar
- Broussard SR, Comuzzie AG, Leighton KL, Leland MM, Whitehead EM, Allan JS. Characterization of new simian foamy viruses from African nonhuman primates. Virology. 1997;237:349–59. DOIPubMedGoogle Scholar
- Herchenroder O, Renne R, Loncar D, Cobb EK, Murthy KK, Schneider J, Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology. 1994;201:187–99. DOIPubMedGoogle Scholar
- Allan JS, Short M, Taylor ME, Su S, Hirsch VM, Johnson PR, Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–28.PubMedGoogle Scholar
- Beer BE, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley SG, Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–44.PubMedGoogle Scholar
- Sharp PM, Bailes E, Gao F, Beer BE, Hirsch VM, Hahn BH. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem Soc Trans. 2000;28:275–82.PubMedGoogle Scholar
- Blewett EL, Black DH, Lerche NW, White G, Eberle R. Simian foamy virus infections in a baboon breeding colony. Virology. 2000;278:183–93. DOIPubMedGoogle Scholar
- Heberling RL, Kalter SS. Isolation of foamy viruses from baboon (Papio cynocephalus) tissues. Am J Epidemiol. 1975;102:25–9.PubMedGoogle Scholar
- Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenroder O, Spatz W, Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology. 1999;257:7–14. DOIPubMedGoogle Scholar
- Lerche NW, Osborn KG, Marx PA, Prahalda S, Maul DH, Lowenstine LJ, Inapparent carriers of simian AIDS type D retrovirus and disease transmission in saliva. J Natl Cancer Inst. 1986;77:489–96.PubMedGoogle Scholar
- Meiering CD, Linial ML. Historical perspective of foamy virus epidemiology and perspective. Clin Microbiol Rev. 2001;14:165–76. DOIPubMedGoogle Scholar
- Khan AS, Sears JF, Muller J, Galvin TA, Shahabuddin M. Sensitive assays for isolation and detection of simian foamy retroviruses. J Clin Microbiol. 1999;37:2678–86.PubMedGoogle Scholar
- Jones-Engel L, Schillaci MA, Engel G. Interaction between humans and nonhuman primates. In Setchell J, Curtis D, editors. Field and laboratory methods in primatology. Cambridge: Cambridge University Press; 2003. p.15–24.
- Jones-Engel L, Engel GA, Schillaci MA. An ethnoprimatological assessment of disease transmission among humans and wild and pet macaques on the Indonesian island of Sulawesi. In: Patterson J, editor. Commensalism and conflict: the primate-human interface. Norman (OK): American Society of Primatologists Publications. In press.
- Engel GA, Jones-Engel L, Schillaci MA, Suaryana KG, Putra A, Fuentes A, Human exposure to herpesvirus B-seropositive macaques, Bali, Indonesia. Emerg Infect Dis. 2002;8:789–95. DOIPubMedGoogle Scholar
- Fuentes A, Southern M, Suaryana KG. Monkey forests and human landscapes: is extensive sympatry sustainable for Homo sapiens and Macaca fascicularis in Bali? In: Patterson J, editor. Commensalism and conflict: the primate-human interface. Norman (OK):American Society of Primatologists Publications. In press.
- Wheatley BP. The sacred monkeys of Bali. Long Grove (IL):Waveland Press; 1998.
- Poinar H, Kuch M, Paabo S. Molecular analysis of oral polio vaccine samples. Science. 2001;292:743–4. DOIPubMedGoogle Scholar
- Vartanian JP, Wain-Hobson S. Analysis of a library of macaque nuclear mitochondrial sequences confirms macaque origin of divergent sequences from old oral polio vaccine samples. Proc Natl Acad Sci U S A. 2002;99:7566–9. DOIPubMedGoogle Scholar
- Sponsel LE. Monkey business? The conservation implications of macaque ethnoprimatology in southern Thailand. In: Fuentes A, Wolfe L, editors. Primates face to face: the conservation implications of human-nonhuman primate interconnections. New York: Cambridge University Press; 2002. p. 288–309.
- Boneva RS, Grindon AJ, Orton SL, Switzer WM, Shanmugam V, Hussain AI, Simian foamy virus infection in a blood donor. Transfusion. 2002;42:886–91. DOIPubMedGoogle Scholar
- Allan JS, Broussard SR, Michaels MG, Starzl TE, Leighton KL, Whitehead EM, Amplification of simian retroviral sequences from human recipients of baboon liver transplants. AIDS Res Hum Retroviruses. 1998;14:821–4. DOIPubMedGoogle Scholar