Volume 11, Number 8—August 2005
Research
Influenza A (H3N2) Outbreak, Nepal
Figure 3
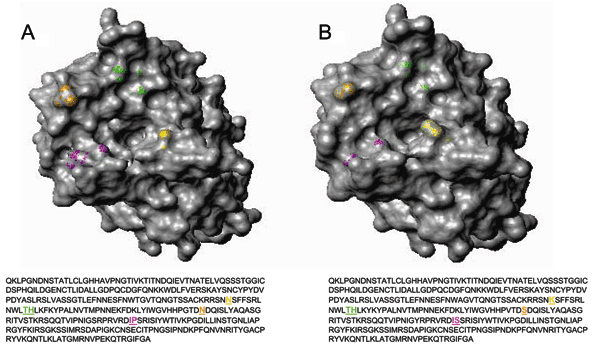
Figure 3. . Three-dimensional top view of the HA1 hemagglutinin structures for A) a representative A/Nepal/1648/04 virus and B) vaccine strain A/Wyoming/3/03. Most (24/26) of the Nepal isolates contain a lysine to asparagine substitution (shown in yellow) at position 145 (K145N). Magenta, residues 226 and 227; orange, residue 189; green, residues 155 and 156; yellow, residue 145. Hemagglutinin molecules were generated by using the respective amino acid sequences with MOLMOL (12). A/Nepal/1648/04 is available from GenBank under accession no. AY945264.
References
- CDC influenza weekly report: influenza summary update, questions and answers, the 2003–2004 influenza season. [cited 19 May 2005]. Available from http://www.cdc.gov/flu/about/qa/fluseason.htm
- Harper SA, Fukuda K, Uyeki TM, Cox NJ, Bridges CB. Centers for Disease Control and Prevention Advisory Committee on Immunization Practices. Prevention and control of influenza: recommendations of the advisory committee on immunization practices (ACIP). MMWR Morb Mortal Wkly Rep. 2004;53:1–40.
- Webster RG, Kendal AP, Gerhard W. Analysis of antigenic drift in recently isolated influenza A (H1N1) viruses using monoclonal antibody preparations. Virology. 1979;96:258–64. DOIPubMedGoogle Scholar
- Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–8. DOIPubMedGoogle Scholar
- Wilson AI, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–87. DOIPubMedGoogle Scholar
- Lee MS, Chen JS. Predicting antigenic variants of influenza A/H3N2 viruses. Emerg Infect Dis. 2004;10:1385–90.PubMedGoogle Scholar
- Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286:1921–5. DOIPubMedGoogle Scholar
- Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc Natl Acad Sci U S A. 2003;100:7152–7. DOIPubMedGoogle Scholar
- Canas LC, Lohman K, Pavlin JA, Endy T, Singh DL, Pandey P, The Department of Defense laboratory-based global influenza surveillance system. Mil Med. 2000;165(Suppl 2):52–6.PubMedGoogle Scholar
- Daum LT, Canas LC, Smith CB, Klimov A, Huff WB, Barnes WJ, Genetic and antigenic analysis of the first A/New Caledonia/20/99-like H1N1 influenza isolates reported in the Americas. Emerg Infect Dis. 2002;8:408–12. DOIPubMedGoogle Scholar
- Kendal AP, Pereura MS, Skehel J. Concepts and procedures for laboratory based influenza surveillance. Geneva: World Health Organization; 1982.
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:29–55, 51–5. DOIPubMedGoogle Scholar
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. •••;18:2714–23. DOIPubMedGoogle Scholar
- Cross KJ, Burleigh LM, Steinhauer DA. Mechanisms of cell entry by influenza virus. Expert Rev Mol Med. 2001;6:1–18.PubMedGoogle Scholar
- Wilson IA, Ladner RC. Skehel, Wiley DC. The structure and role of the carbohydrate moieties of influenza virus hemagglutinin. Biochem Soc Trans. 1983;11:145–7.
- Hughey PG, Roberts PC, Holsinger LJ, Zebedee SL, Lamb RA, Compans RW. Effects of antibody to the influenza A virus M2 protein on M2 surface expression and virus assembly. Virology. 1995;212:411–21. DOIPubMedGoogle Scholar
- Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–72.PubMedGoogle Scholar
- Webster RG, Brown LE, Laver WG. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology. 1984;135:30–42. DOIPubMedGoogle Scholar
- Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–8.PubMedGoogle Scholar
- CDC weekly report: influenza summary update (week ending April 16, 2005). [cited 19 May 2005]. Available from http://www.cdc.gov/flu/weekly/
Page created: April 23, 2012
Page updated: April 23, 2012
Page reviewed: April 23, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.