Genome Analysis Linking Recent European and African Influenza (H5N1) Viruses
Steven L. Salzberg*

, Carl Kingsford*, Giovanni Cattoli†, David J. Spiro‡, Daniel A. Janies§, Mona Mehrez Aly¶, Ian H. Brown#, Emmanuel Couacy-Hymann**, Gian Mario De Mia††, Do Huu Dung‡‡, Annalisa Guercio§§, Tony Joannis¶¶, Ali Safar Maken Ali##, Azizullah Osmani***, Iolanda Padalino†††, Magdi D. Saad‡‡‡, Vladimir Savi槧§, Naomi A. Sengamalay‡, Samuel L. Yingst‡‡‡, Jennifer Zaborsky‡, Olga Zorman-Rojs¶¶¶, Elodie Ghedin###, and Ilaria Capua†
Author affiliations: *University of Maryland Center for Bioinformatics and Computational Biology, College Park, Maryland, USA; †Istituto Zooprofilattico Sperimentale delle Venezie, Padova, Italy; ‡The Institute for Genomic Research, Rockville, Maryland, USA; §Ohio State University, Columbus, Ohio, USA; ¶Animal Health Research Institute, Giza, Egypt; #Veterinary Laboratories Agency, Addlestone, England, UK; **Central Laboratory of Animal Pathology, Bingerville, Côte d’Ivoire; ††Istituto Zooprofilattico Sperimentale dell’Umbria e delle Marche, Perugia, Italy; ‡‡Department of Animal Health, Hanoi, Vietnam; §§Istituto Zooprofilattico Sperimentale della Sicilia, Palermo, Italy; ¶¶National Veterinary Research Institute, Vom. Plateau State, Nigeria; ##Food and Agriculture Office of the United Nations, Tehran, Iran; ***Ministry of Agriculture, Animal Husbandry and Food, Kabul, Afghanistan; †††Istituto Zooprofilattico Sperimentale della Puglia e Basilicata, Foggia, Italy; ‡‡‡US Naval Medical Research Unit No. 3, Cairo, Egypt; §§§Croatian Veterinary Institute, Zagreb, Croatia; ¶¶¶University of Ljubljana, Ljubljana, Slovenia; ###University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA;
Main Article
Figure 2
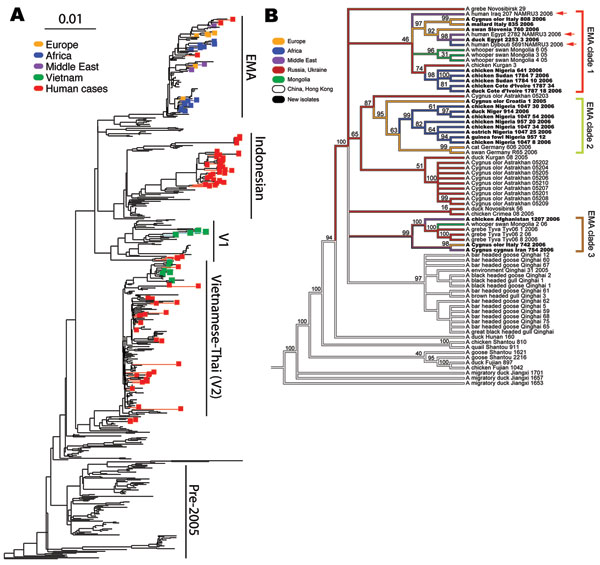
Figure 2. A) Phylogenetic tree relating the influenza A (H5N1) hemagglutinin (HA) segments of 589 avian, feline, and human viruses. The tree includes all HA segments isolated since 2000 from humans (82 isolates, minimum sequence length 1,000 nt), birds (503 isolates, minimum length 1500 nt), and cats (4 isolates). The 36 newly sequenced genomes are highlighted in color. Human cases, which occur in all 4 of the major influenza (H5N1) clades, are highlighted in red. The scale bar indicates an F84 distance of 0.01. A full-scale version of this tree is provided as Figure 3. B) Phylogeny of 71 complete genomes (avian isolates, all 8 segments concatenated) and 3 HA sequences (human isolates, marked with red arrows) from Europe, the Middle East, Africa, Russia, and Asia. Bootstrap values represent the percentage of 1,000 bootstrap replicates for which the partition implied by the edge was observed; see Methods for further details. The 3 European-Middle Eastern-African (EMA) subclades from Figure 1 are indicated with the same color scheme. Isolates from human hosts are found only in EMA-1. Colors indicate locales. The names of the isolates newly sequenced in this study are shown in boldface text.
Main Article
Page created: June 24, 2010
Page updated: June 24, 2010
Page reviewed: June 24, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
