Volume 16, Number 5—May 2010
Dispatch
Nosocomial Outbreak of Crimean-Congo Hemorrhagic Fever, Sudan
Figure
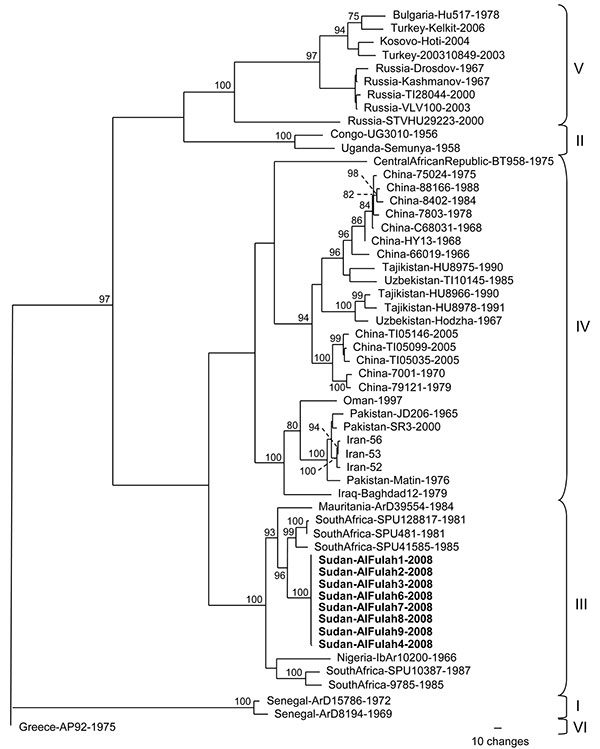
Figure. Phylogenetic relationship of Crimean-Congo hemorrhagic fever virus (CCHFV) full-length small (S) segments. Phylogenetic analysis used 47 full-length CCHFV S segments available in GenBank. GARLI (v0.96b8) (9) with default settings was used to generate a maximum-likelihood tree with bootstrap support values from 1,000 replicates. From the analysis, a 50% majority-rule tree was constructed. Virus strains from Sudan patients 1, 2, 3, 6, 7, 8, and 9 (GenBank accession no. GQ862371) were identical, and the virus sequence from patient 4 (GenBank accession no. GQ862372) differed by 1 nt. Each strain is listed by its location, strain name, and year of isolation, when available. Boldface indicates strains from Sudan; braces indicate previously described genetic lineages (8).
References
- Hoogstraal H. The epidemiology of tick-borne Crimean-Congo haemorrhagic fever in Asia, Europe and Africa. J Med Entomol. 1979;15:307–417.PubMedGoogle Scholar
- Swanepoel R. Crimean-Congo haemorrhagic fever. In: Coetzer JA, Thomson GR, Tustin RC, editors. Infectious diseases of livestock with special reference to southern Africa. Cape Town (South Africa): Oxford University Press; 1994. p. 723–9.
- Whitehouse CA. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004;64:145–60. DOIPubMedGoogle Scholar
- Ergonul O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6:203–14. DOIPubMedGoogle Scholar
- Schmaljohn CS, Nichol ST. Bunyaviridae. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, et al., editors. Fields virology, 5th ed. Philadelphia: Lippincott, Williams, and Wilkins; 2007. p. 1741–89.
- Burney MI, Ghafoor A, Saleen M, Webb PA, Casals J. Nosocomial outbreak of viral hemorrhagic fever caused by Crimean hemorrhagic fever–Congo virus in Pakistan, January 1976. Am J Trop Med Hyg. 1980;29:941–7.PubMedGoogle Scholar
- Schwarz TF, Nsanze H, Longson M, Nitschko H, Gilch S, Shurie H, Polymerase chain reaction for diagnosis and identification of distinct variants of Crimean-Congo hemorrhagic fever virus in the United Arab Emirates.Am J Trop Med Hyg. 1996;55:190–6.
- Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. 2006;80:8834–42. DOIPubMedGoogle Scholar
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum-likelihood criterion [dissertation]. Austin (TX): The University of Texas; 2006.
- Watts DM, Tigani AE, Botros BA, Salib AW, Olson JG, McCarthy M, Arthropod-borne viral infections associated with a fever outbreak in the northern province of Sudan.J Trop Med Hyg. 1994;97:228–30.
- McCarthy MC, Haberberger RL, Salib AW, Soliman BA, Tigani AE, Khalid OI, Evaluation of arthropod-borne viruses and other infectious disease pathogens as the causes of febrile illnesses in the Khartoum Province of Sudan.J Med Virol. 1996;48:141–6.
- Morrill JC, Soliman AK, Imam IZ, Botros BA, Moussa MI, Watts DM. Serological evidence of Crimean-Congo haemorrhagic fever viral infection among camels imported into Egypt. J Trop Med Hyg. 1990;93:201–4.PubMedGoogle Scholar
- Hassanein KM, el-Azazy A, Yousef HM. Trans R Soc Trop Med Hyg. 1997;91:536–7.Detection of Crimean-Congo haemorrhagic fever virus antibodies in humans and imported livestock in Saudi Arabia. DOIPubMedGoogle Scholar
- Chamberlain J, Cook N, Lloyd G, Mioulet V, Tolley H, Hewson R. Co-evolutionary patterns of variation in small and large RNA segments of Crimean-Congo hemorrhagic fever virus. J Gen Virol. 2005;86:3337–41. DOIPubMedGoogle Scholar
- Burt FJ, Paweska JT, Ashkettle B, Swanepoel R. Genetic relationship in southern African Crimean-Congo haemorrhagic fever virus isolates: evidence for occurrence of reassortment. Epidemiol Infect. 2009;137:1302–8. DOIPubMedGoogle Scholar