Volume 16, Number 9—September 2010
Research
Worldwide Diversity of Klebsiella pneumoniae That Produce β-Lactamase blaKPC-2 Gene1
Figure 1
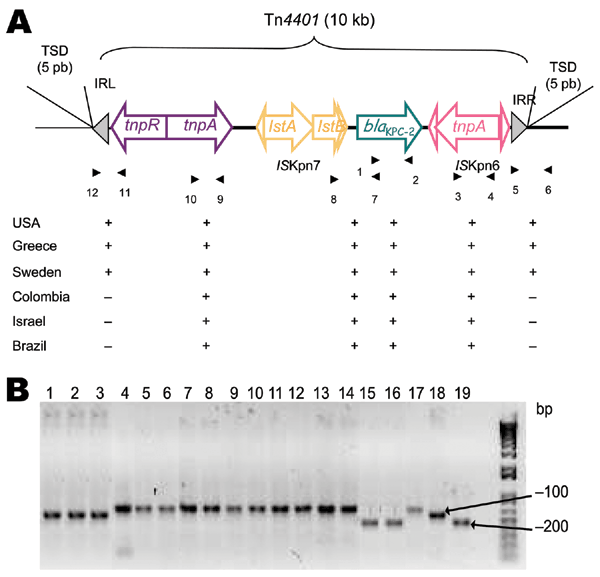
Figure 1. A) Schematic representation of Tn4401 isoforms on plamids of Klebsiella pneumoniae isolates that produce K. pneumoniae carbapenemases (KPCs). Genes and their corresponding transcription orientations are indicated by horizontal arrows. Gray triangles represent the inverted repeats left (IRL) and right (IRR) of Tn4401. Small and empty triangles represent the inverted repeats of ISKpn6 and ISKpn7. Target site duplications (TSD) are indicated above the sequence. Primers listed in Table 2 are shown below, with results of PCRs for each isolate. B) PCR results with primers 7 and 8 (Table 2). Lane 1, K. pneumoniae YC (11); lane 2, K. pneumoniae GR (21); lane 3, K. pneumoniae K271 (25); lane 4, K. pneumoniae KN2303 (13); lane 5, K. pneumoniae KN633 (13); lane 6, K. pneumoniae INC H1521-6; lane 7, K. pneumoniae INC H1516-6; lane 8, K. pneumoniae HPTU 27635; lane 9, K. pneumoniae HPTU 2020532; lane 10, K. pneumoniae A28006 (16); lane 11, K. pneumoniae A28008 (16); lane 12, K. pneumoniae A28009 (16); lane 13, K. pneumoniae A28011 (16); lane 14, K. pneumoniae A33504 (16); lane 15, K. pneumoniae 475; lane 16, K. pneumoniae 588.
References
- Nordmann P, Poirel L. Emerging carbapenemases in gram-negatives aerobes. Clin Microbiol Infect. 2002;8:321–31. DOIPubMedGoogle Scholar
- Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58. DOIPubMedGoogle Scholar
- Poirel L, Héritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. DOIPubMedGoogle Scholar
- Nordmann P, Cuzon G, Naas T. The real threat of KPC carbapenemase–producing bacteria. Lancet Infect Dis. 2009;9:228–36. DOIPubMedGoogle Scholar
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Novel carbapenem-hydrolyzing β-lactamase KPC-1 from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. DOIPubMedGoogle Scholar
- Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis. 2004;39:55–60. DOIPubMedGoogle Scholar
- Landman D, Bratu S, Kochar S, Panwar M, Trehan M, Doymaz M, Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. J Antimicrob Chemother. 2007;60:78–82. DOIPubMedGoogle Scholar
- Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Molecular epidemiology of KPC-producing Klebsiella pneumoniae in the United States: clonal expansion of MLST sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70. DOIPubMedGoogle Scholar
- Miriagou V, Tzouvelekis LS, Rossiter S, Tzelepi E, Angulo FJ, Whichard J. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob Agents Chemother. 2003;47:1297–300. DOIPubMedGoogle Scholar
- Wolter DJ, Khalaf N, Robledo IE, Vazquez GJ, Sante MI, Aquino EE, Surveillance of carbapenem-resistant Pseudomonas aeruginosa from Puerto Rico medical center hospitals: dissemination of KPC and IMP-18 beta-lactamases. Antimicrob Agents Chemother. 2009;53:1660–4. DOIPubMedGoogle Scholar
- Naas T, Nordmann P, Vedel G, Poyart C. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob Agents Chemother. 2005;49:4423–4. DOIPubMedGoogle Scholar
- Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob Agents Chemother. 2007;51:3026–9. DOIPubMedGoogle Scholar
- Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, Vallejo M; Colombian Nosocomial Resistance Study Group. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother. 2006;50:2880–2. DOIPubMedGoogle Scholar
- Monteiro J, Fernandes Santos A, Asensi MD, Peirano G, Gales AC. First report of KPC-2–producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53:333–4. DOIPubMedGoogle Scholar
- Pasteran FG, Otaegui L, Guerriero L, Radice G, Maggiora R, Rapoport M, Klebsiella pneumoniae carbapenemase-2, Buenos Aires, Argentina. Emerg Infect Dis. 2008;14:1178–80. DOIPubMedGoogle Scholar
- Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP; Colombian Nosocomial Resistance Study Group. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother. 2007;51:1553–5. DOIPubMedGoogle Scholar
- Cai JC, Zhou HW, Zhang R, Chen GX. Emergence of Serratia marscescens, Klebsiella pneumoniae, and Escherichia coli possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units from a Chinese hospital. Antimicrob Agents Chemother. 2008;52:2014–8. DOIPubMedGoogle Scholar
- Pournaras S, Protonotariou E, Voulgari E, Kristo I, Dimitroulia E, Vitti D, Clonal spread of KPC-2 carbapenemase-producing Klebsiella pneumoniae strains in Greece. J Antimicrob Chemother. 2009;64:348–52. DOIPubMedGoogle Scholar
- Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, First report on hyper-epidemic clone of KPC-3 producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother. 2009;53:818–20. DOIPubMedGoogle Scholar
- Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, Macdonald A, Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J Antimicrob Chemother. 2008;62:1261–4. DOIPubMedGoogle Scholar
- Cuzon G, Naas T, Demachy MC, Nordmann P. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from Greece. Antimicrob Agents Chemother. 2008;52:796–7. DOIPubMedGoogle Scholar
- Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother. 2008;52:1257–63. DOIPubMedGoogle Scholar
- Wolter DJ, Kurpiel PM, Woodford N, Palepou MF, Goering RV, Hanson ND. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob Agents Chemother. 2009;53:557–62. DOIPubMedGoogle Scholar
- Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Novel genetic environment of the carbapenem-hydrolysing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53:4333–8. DOIPubMedGoogle Scholar
- Tegmark Wisell K, Haeggman S, Gazelius L, Thompson O, Gustafsson I, Ripa T, Identification of Klebsiella pneumoniae carbapenemase in Sweden. Euro Surveill. 2007;12:E071220.3.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. M100–S15. Wayne (PA): The Institute; 2005.
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. DOIPubMedGoogle Scholar
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989.
- Carrër A, Lassel L, Fortineau N, Mansouri M, Anguel N, Richard C, Outbreak of CTX-M-15–producing Klebsiella pneumoniae in the intensive care unit of a French hospital. Microb Drug Resist. 2009;15:47–54. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit R, Goering V, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–82. DOIPubMedGoogle Scholar
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. DOIPubMedGoogle Scholar
- Falagas ME, Rafailidis PI, Ioannidou E, Alexiou VG, Matthaiou DK, Karageorgopoulos DE, Colistin therapy for microbiologically documented multidrug-resistant gram-negative bacterial infections: a retrospective cohort study of 258 patients. Int J Antimicrob Agents. 2010;35:194–9. Epub 2009 Dec 16. DOIPubMedGoogle Scholar
- Hæggman S, Löfdahl S, Paauw A, Verhoef J, Brisse S. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:2400–8. DOIPubMedGoogle Scholar
- Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother. 2009;53:1998–2004. DOIPubMedGoogle Scholar
- Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother. 2009;63:654–8. DOIPubMedGoogle Scholar
- Baraniak A, Izdebski R, Herda M, Fiett J, Hryniewicz W, Gniadkowski M. The emergence of Klebsiella pneumoniae ST258 with KPC-2 in Poland. Antimicrob Agents Chemother. 2009;53:4565–7. DOIPubMedGoogle Scholar
- Damjanova I, Toth A, Paszti J, Hajbel-Vekony G, Jakab M, Berta J, Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15–type beta-lactamase–producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new “MRSAs”? J Antimicrob Chemother. 2008;62:978–85. DOIPubMedGoogle Scholar
- Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J Antimicrob Chemother. 2009;63:427–37. DOIPubMedGoogle Scholar
- Willems RJ, Top J, Van den Braak N, Van Belkum A, Mevius DJ, Hendriks G, Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999;43:483–91.PubMedGoogle Scholar
1This study was presented in part at the Interscience Conference on Antimicrobial Agents and Chemotherapy; 2009 Sep 12–15; San Francisco, CA, USA.