Volume 17, Number 1—January 2011
Research
Genotyping Rotavirus RNA from Archived Rotavirus-Positive Rapid Test Strips
Figure
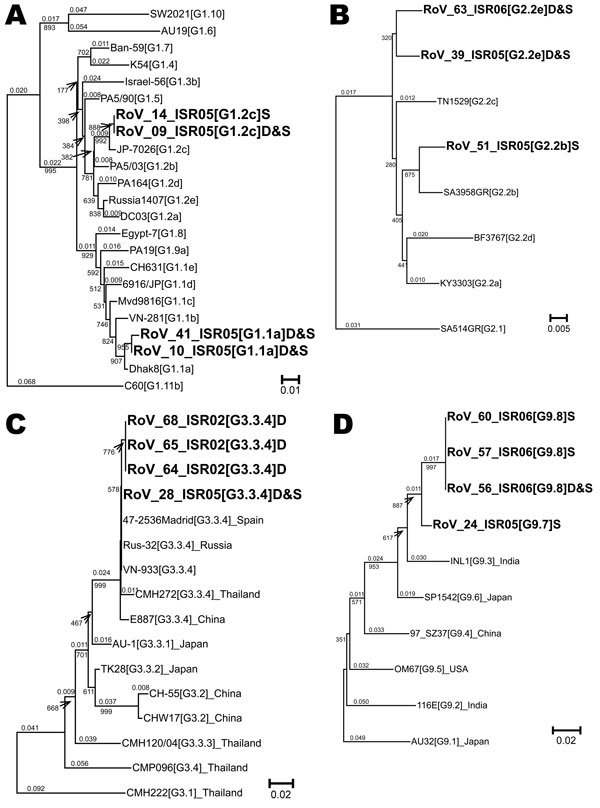
Figure. Neighbor-joining phylogenetic trees for viral protein (VP) 7 G1, G2, G3, and G9 genotypes of hospitalized children in Israel, including sequences recovered from archived rotavirus dipsticks. Representative isolates for lineages and sublineages of VP7 genotypes G1 (A), G2 (B), G3 (C), and G9 (D) were chosen from Phan et al. (15) Page and Steele (16), Wang et al. (17), and Martinez-Laso et al. (18), respectively, and the sequences were downloaded from the European Molecular Biology Laboratory/GenBank/DNA Data Bank of Japan. These sequences were aligned with Israeli sequences by using the Sequencher program (Genecodes, Ann Arbor, MI, USA) and truncated to the longest segment common to all sequences in the alignment; 515 nt for G1, 547 nt for G2, 274 nt for G3, and 207 nt for G9. Each of the 4 phylogenetic trees was prepared by using ClustalX (13) for data bootstrapped 1,000× and was analyzed with NJplot (14). Whole numbers indicate bootstrap values for branches; fractional numbers indicate genetic distances. The genotype, lineage, and, where relevant, sublineage of each isolate appears in brackets after the name of the isolate: for example, KY3303[G2.2a] is VP7 genotype G 2, lineage 2, sublineage a for isolate KY3303. A letter at the end of the name of the Israeli sequences indicates the source of the RNA (D for dipstick or S for fecal suspension). D&S appears when the sequences were identical. The GenBank accession numbers for all sequences in this figure appear in the online Appendix Table (Table A). Scale bars indicate percent of nucleotide substitutions per site.
References
- Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis. 2003;9:565–72.PubMedGoogle Scholar
- Muhsen K, Shulman L, Rubinstein U, Kasem E, Kremer A, Goren S, Incidence, characteristics, and economic burden of rotavirus gastroenteritis associated with hospitalization of Israeli children <5 years of age, 2007–2008. J Infect Dis. 2009;200(Suppl 1):S254–63. DOIPubMedGoogle Scholar
- Widdowson MA, Bresee JS, Gentsch JR, Glass RI. Rotavirus disease and its prevention. Curr Opin Gastroenterol. 2005;21:26–31.PubMedGoogle Scholar
- Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12:304–6.PubMedGoogle Scholar
- Parashar UD, Burton A, Lanata C, Boschi-Pinto C, Shibuya K, Steele D, Global mortality associated with rotavirus disease among children in 2004. J Infect Dis. 2009;200(Suppl 1):S9–15. DOIPubMedGoogle Scholar
- Estes MK, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–49.PubMedGoogle Scholar
- Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, et al., editors. Fields virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. p. 1917–74.
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Safety and efficacy of a pentavalent human–bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23–33. DOIPubMedGoogle Scholar
- Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, Clemens SC, Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11–22. DOIPubMedGoogle Scholar
- Muhsen K, Shulman L, Kasem E, Rubinstein U, Shachter J, Kremer A, Effectiveness of rotavirus vaccines for prevention of rotavirus gastroenteritis–associated hospitalizations in Israel: a case–control study. [Epub ahead of print.]. Hum Vaccin. 2010;6.PubMedGoogle Scholar
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–82.PubMedGoogle Scholar
- Gunasena S, Nakagomi O, Isegawa Y, Kaga E, Nakagomi T, Steele AD, Relative frequency of VP4 gene alleles among human rotaviruses recovered over a 10-year period (1982–1991) from Japanese children with diarrhea. J Clin Microbiol. 1993;31:2195–7.PubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. DOIPubMedGoogle Scholar
- Perrière G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–9. DOIPubMedGoogle Scholar
- Phan TG, Khamrin P, Quang TD, Dey SK, Takanashi S, Okitsu S, Detection and genetic characterization of group A rotavirus strains circulating among children with acute gastroenteritis in Japan. J Virol. 2007;81:4645–53. DOIPubMedGoogle Scholar
- Page NA, Steele AD. Antigenic and genetic characterization of serotype G2 human rotavirus strains from the African continent. J Clin Microbiol. 2004;42:595–600. DOIPubMedGoogle Scholar
- Wang YH, Kobayashi N, Zhou X, Nagashima S, Zhu ZR, Peng JS, Phylogenetic analysis of rotaviruses with predominant G3 and emerging G9 genotypes from adults and children in Wuhan, China. J Med Virol. 2009;81:382–9. DOIPubMedGoogle Scholar
- Martinez-Laso J, Roman A, Head J, Cervera I, Rodriguez M, Rodriguez-Avial I, Phylogeny of G9 rotavirus genotype: a possible explanation of its origin and evolution. J Clin Virol. 2009;44:52–7. DOIPubMedGoogle Scholar
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. DOIPubMedGoogle Scholar
- Silberstein I, Shulman LM, Mendelson E, Shif I. Distribution of both rotavirus VP4 genotypes and VP7 serotypes among hospitalized and nonhospitalized Israeli children. J Clin Microbiol. 1995;33:1421–2.PubMedGoogle Scholar
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. DOIPubMedGoogle Scholar
- Santos N, Volotao EM, Soares CC, Campos GS, Sardi SI, Hoshino Y. Predominance of rotavirus genotype G9 during the 1999, 2000, and 2002 seasons among hospitalized children in the city of Salvador, Bahia, Brazil: implications for future vaccine strategies. J Clin Microbiol. 2005;43:4064–9. DOIPubMedGoogle Scholar