Volume 17, Number 4—April 2011
Research
Genomic Analysis of Highly Virulent Georgia 2007/1 Isolate of African Swine Fever Virus
Figure 2
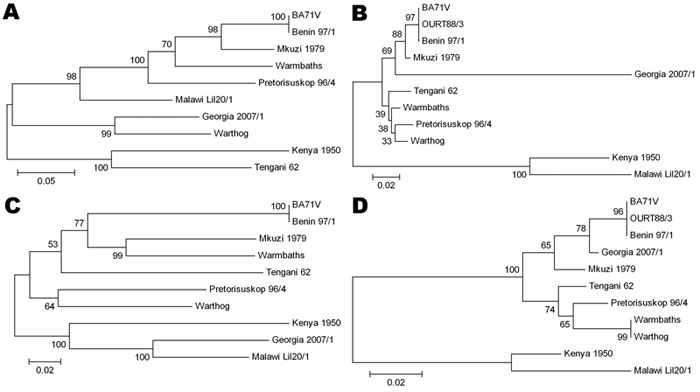
Figure 2. Phylogenetic trees of 4 of the most divergent African swine fever virus proteins. A) C-type lectin EP153R, B) A238L, C) CD2-like protein EP402R, D) structural protein K177R (P22). Evolutionary history was inferred by using the neighbor-joining method. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the proteins analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated proteins clustered in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were 224 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (27). Scale bars indicate amino acid substitutions per site.
References
- Costard S, Wieland B, de Glanville W, Jori F, Rowlands R, Vosloo W, African swine fever: how can global spread be prevented? Philos Trans R Soc Lond B Biol Sci. 2009;364:2683–96. DOIPubMedGoogle Scholar
- Dixon LK, Escribano JM, Martins C, Rock DL, Salas ML, Wilkinson PJ. Asfarviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2005. p. 135–43.
- Rojo G, Garcia-Beato R, Vinuela E, Salas ML, Salas J. Replication of African swine fever virus DNA in infected cells. Virology. 1999;257:524–36. DOIPubMedGoogle Scholar
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–84. DOIPubMedGoogle Scholar
- ProMED-mail. African swine fever—Russia (05): (KB), conf., culled. 2010 Apr 6 [cited 2011 Jan 6]. http://www.promedmail.org, archive no. 20100406.1107.
- Bastos ADS, Penrith ML, Cruciere C, Edrich JL, Hutchings G, Roger F, Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch Virol. 2003;148:693–706. DOIPubMedGoogle Scholar
- Rowlands RJ, Michaud V, Heath L, Hutchings G, Oura C, Vosloo W, African swine fever virus isolate, Georgia, 2007. Emerg Infect Dis. 2008;14:1870–4. DOIPubMedGoogle Scholar
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. DOIPubMedGoogle Scholar
- Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–8. DOIPubMedGoogle Scholar
- Tcherepanov V, Ehlers A, Upton C. Genome annotation transfer utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics. 2006;7:150. DOIPubMedGoogle Scholar
- Upton C, Hogg D, Perrin D, Boone M, Harris NL. Viral genome organizer: a system for analyzing complete viral genomes. Virus Res. 2000;70:55–64. DOIPubMedGoogle Scholar
- Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. DOIPubMedGoogle Scholar
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363. DOIPubMedGoogle Scholar
- Chapman DAG, Tcherepanov V, Upton C, Dixon LK. Comparison of the genome sequences of nonpathogenic and pathogenic African swine fever virus isolates. J Gen Virol. 2008;89:397–408. DOIPubMedGoogle Scholar
- Yáñez RJ, Rodriguez JM, Nogal ML, Yuste L, Enriquez C, Rodriguez JF, Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–78. DOIPubMedGoogle Scholar
- Yozawa T, Kutish GF, Afonso CL, Lu Z, Rock DL. Two novel multigene families, 530 and 300, in the terminal variable regions of African swine fever virus genome. Virology. 1994;202:997–1002. DOIPubMedGoogle Scholar
- Boinas FS, Hutchings GH, Dixon LK, Wilkinson PJ. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J Gen Virol. 2004;85:2177–87. DOIPubMedGoogle Scholar
- Zsak L, Borca MV, Risatti GR, Zsak A, French RA, Lu Z, Preclinical diagnosis of African swine fever in contact-exposed swine by a real-time PCR assay. J Clin Microbiol. 2005;43:112–9. DOIPubMedGoogle Scholar
- Haresnape JM, Wilkinson PJ. A study of African swine fever virus–infected ticks (Ornithodoros moubata) collected from 3 villages in the ASF enzootic area of Malawi following an outbreak of the disease in domestic pigs. Epidemiol Infect. 1989;102:507–22. DOIPubMedGoogle Scholar
- Pan IC. African swine fever virus: generation of subpopulations with altered immunogenicity and virulence following passage in cell cultures. J Vet Med Sci. 1992;54:43–52.PubMedGoogle Scholar
- Borca MV, Carrillo C, Zsak L, Laegreid WW, Kutish GF, Neilan JG, Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J Virol. 1998;72:2881–9.PubMedGoogle Scholar
- Hurtado C, Granja AG, Bustos MJ, Nogal ML, de Buitrago GG, de Yebenes VG, The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology. 2004;326:160–70. DOIPubMedGoogle Scholar
- Goatley LC, Marron MB, Jacobs SC, Hammond JM, Miskin JE, Abrams CC, Nuclear and nucleolar localization of an African swine fever virus protein, I14L, that is similar to the herpes simplex virus-encoded virulence factor ICP34.5. J Gen Virol. 1999;80:525–35.PubMedGoogle Scholar
- Sussman MD, Lu Z, Kutish G, Afonso CL, Roberts P, Rock DL. Identification of an African swine fever virus gene with similarity to a myeloid differentiation primary response gene and a neurovirulence-associated gene of herpes simplex virus. J Virol. 1992;66:5586–9.PubMedGoogle Scholar
- Zsak L, Lu Z, Kutish GF, Neilan JG, Rock DL. An African swine fever virus virulence-associated gene NL-S with similarity to the herpes simplex virus ICP34.5 gene. J Virol. 1996;70:8865–71.PubMedGoogle Scholar
- Camacho A, Vinuela E. Protein p22 of African swine fever virus: an early structural protein that is incorporated into the membrane of infected cells. Virology. 1991;181:251–7. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Neilan JG, Zsak L, Lu Z, Kutish GF, Afonso CL, Rock DL. Novel swine virulence determinant in the left variable region of the African swine fever virus genome. J Virol. 2002;76:3095–104. DOIPubMedGoogle Scholar
- de Villiers EP, Gallardo C, Arias M, da Silva M, Upton C, Martin R, Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology. 2010;400:128–36. DOIPubMedGoogle Scholar