Volume 17, Number 4—April 2011
Letter
Sequence Analysis of Feline Coronaviruses and the Circulating Virulent/Avirulent Theory
Figure
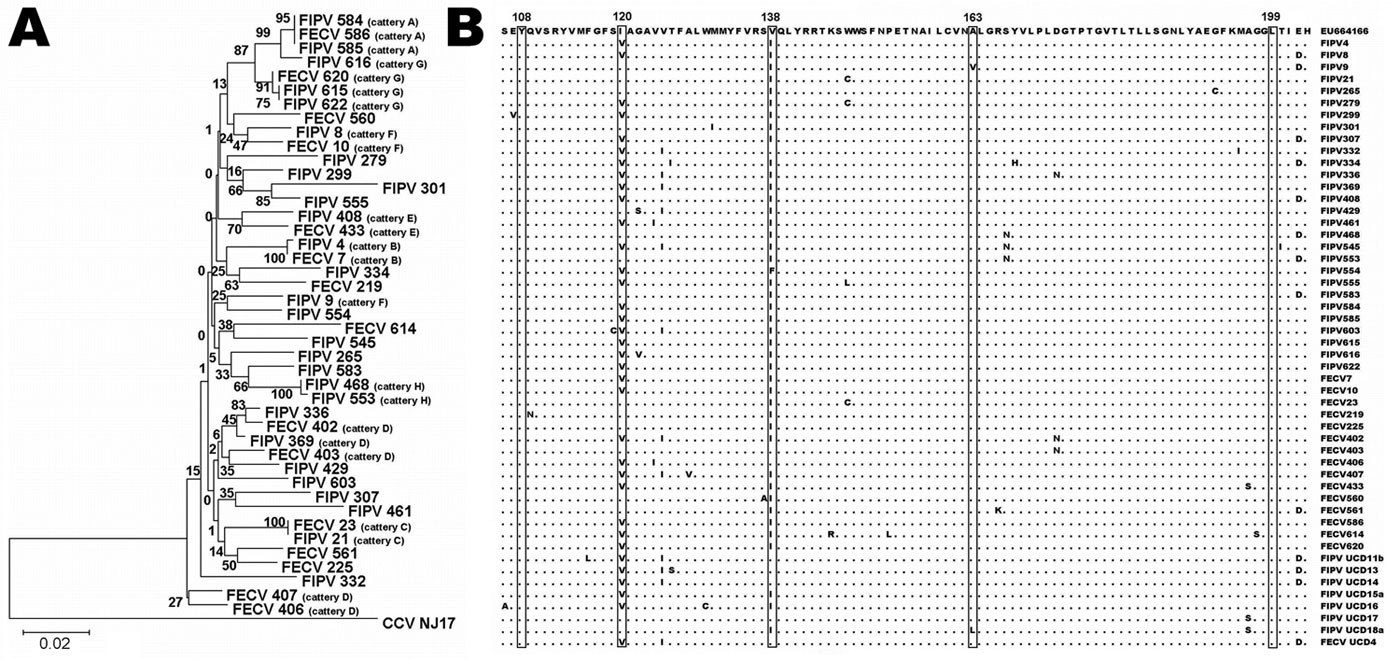
Figure. A) Phylogenetic relationships of feline coronaviruses (FCoVs) detected in feces of healthy cats and in organs/ascites of cats with feline infectious peritonitis. Alignment of the matrix (M) gene sequences was used to generate a rooted neighbor-joining tree with the M gene sequence of canine coronavirus strain NJ17 (Genbank accession no. AY704917) as outgroup. Bootstrap confidence values (percentages of 1,000 replicates) are indicated at the relevant branching points. Branch lengths are drawn to scale; scale bar indicates 0.02 nucleotide substitutions per site. Viruses detected in cattery animals are indicated by a cattery designation after the virus identification number. B) Alignment of amino acid sequences of partial M proteins of the FCoVs from panel A, as compared with a feline infectious peritonitis virus (FIPV) reference sequence (top line) published by Brown et al. (9) (GenBank accession no. EU664166), and with 8 American FCoV sequences (bottom) published by Pedersen et al. (8). The 5 aa residues at positions 108, 120, 138, 163, and 199, suggested by Brown et al. (9) as potential diagnostic sites, are boxed.
References
- Addie D, Belák S, Boucraut-Baralon C, Egberink H, Frymus T, Gruffydd-Jones T, Feline infectious peritonitis. ABCD guidelines on prevention and management. J Feline Med Surg. 2009;11:594–604. DOIPubMedGoogle Scholar
- de Groot RJ, Horzinek MC. Feline infectious peritonitis. In: The Coronaviridae. Siddell SG, editor. New York: Plenum Press; 1995. p. 293–309.
- Haijema BJ, Rottier PJ, de Groot RJ. Feline coronaviruses: a tale of two-faced types. In: Thiel V, editor. Coronaviruses. Molecular and cellular biology. Norfolk (UK): Academic Press; 2007. p. 183–203.
- Pedersen NC. A review of feline infectious peritonitis virus infection: 1963-2008. J Feline Med Surg. 2009;11:225–58. DOIPubMedGoogle Scholar
- Herrewegh AA, Vennema H, Horzinek MC, Rottier PJM, de Groot RJ. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212:622–31. DOIPubMedGoogle Scholar
- Poland AM, Vennema H, Foley JE, Pedersen NC. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J Clin Microbiol. 1996;34:3180–4.PubMedGoogle Scholar
- Vennema H, Poland A, Foley J, Pedersen NC. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–7. DOIPubMedGoogle Scholar
- Pedersen NC, Liu H, Dodd KA, Pesavento PA. Significance of coronavirus mutants in feces and diseased tissues of cats suffering from feline infectious peritonitis. Viruses. 2009;1:166–84. DOIGoogle Scholar
- Brown MA, Troyer JL, Pecon-Slattery J, Roelke ME, O’Brien SJ. Genetics and pathogenesis of feline infectious peritonitis virus. Emerg Infect Dis. 2009;15:1445–52. DOIPubMedGoogle Scholar
- Chang HW, de Groot RJ, Egberink HF, Rottier PJ. Feline infectious peritonitis: insights into feline coronavirus pathobiogenesis and epidemiology based on genetic analysis of the viral 3c gene. J Gen Virol. 2010;91:415–20. DOIPubMedGoogle Scholar