Volume 17, Number 5—May 2011
Letter
Fatal Human Case of Western Equine Encephalitis, Uruguay
Figure
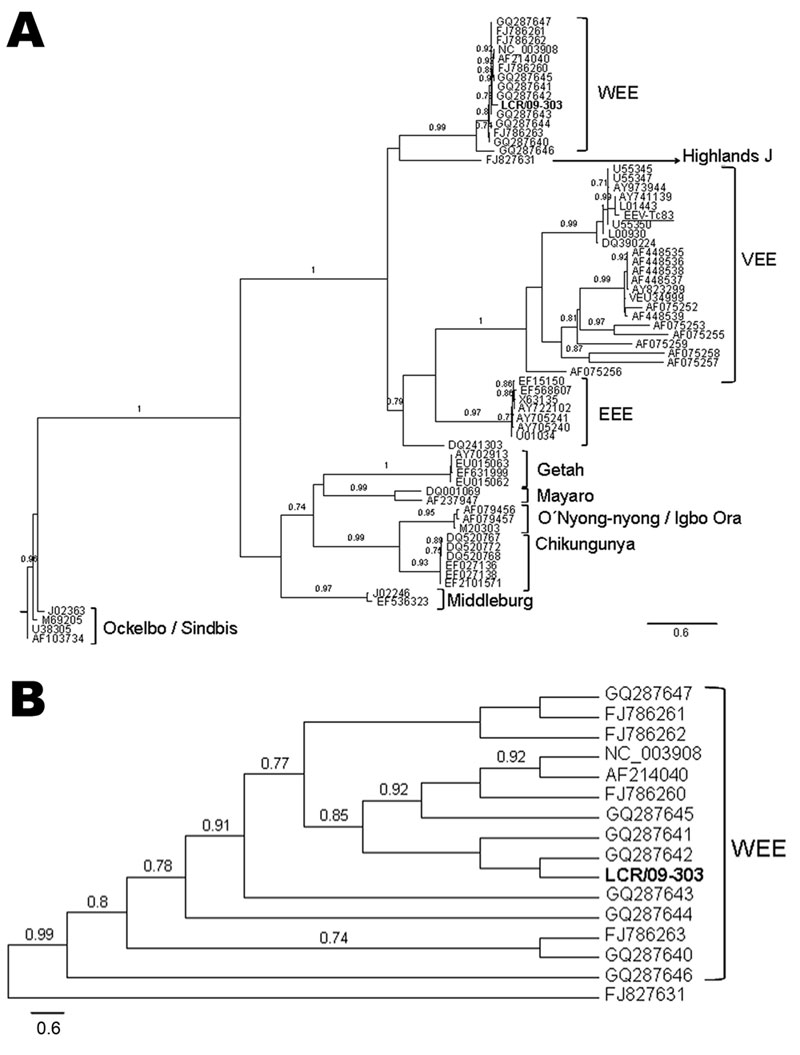
Figure. A) Phylogenetic tree obtained by maximum likelihood analysis of sequences corresponding to the alphavirus NSP4 gene. Alignment used in the analysis had 448 bp and was conducted by using BioEdit software version 7.0.9.0 (www.mbio.ncsu.edu/BioEdit/BioEdit.html). Estimation of the suitable model of nucleotide substitution was carried out by using Modelgenerator (http://bioinf.may.ie/software/modelgenerator). Phylogenetic analysis was run on the PhyML web server (www.atgc-montpellier.fr/phyml), with the following settings: nucleotide substitution model: general time reversible + proportion invariant + Γ; proportion of invariable sites: 0.39; gamma distribution parameter α: 0.67; node support: approximate likelihood-ratio test (only values over 0.70 are shown). Sequences included in the analysis were the following (GenBank accession numbers for individual isolates provided when applicable): human encephalitis case-patient: LCR/09-303 (boldface); reverse transcription–PCR positive control Venezuelan equine encephalitis virus [VEEV] Tc83 (282 nt), FJ786261; western equine encephalitis virus (WEEV): AF214040, GQ287647, GQ287646, GQ287645, GQ287644, GQ287643, GQ287642, GQ287641, GQ287640, FJ786263, FJ786262, FJ786260, NC003908; Highlands J virus, FJ827831; Venezuelan equine encephalitis virus (VEEV), L01443, DQ390224, AF075255, AY823299, AF448539, AF448538, AF448537, AF448536, AF448535, AF075252, U34999, AF075259, AF075256, AF075253, AF075257, AF075258, AY973944. L00930, AY741139, U55350, U55347, U55345; eastern equine encephalitis virus (EEEV), AY722102, U01034, EF568607, EF15150, AY705241, AY705240, X63135, DQ241303; Getah virus, EU015063, EU015062, EF631999, AY702913; Mayaro virus, AF237947, DQ001069; M20303; O’nyong-nyong virus, AF079456; Igbo Ora virus, AF079457; chikungunya virus, EF210157, EF027138, EF027136, DQ520772, DQ520768, DQ520767; Middleburg virus, EF536323, J02246; Ockelbo virus, M69205: Sindbis virus, AF103734, U38305, J02363, M69205. B) Detail of the WEEV clade, showing the relationships between the sample LCR/09-303 and the rest of the WEEV isolates included in the analysis. Scale bars indicate expected nucleotide changes per site.