Volume 17, Number 5—May 2011
Dispatch
Novel Bluetongue Virus Serotype from Kuwait
Figure 2
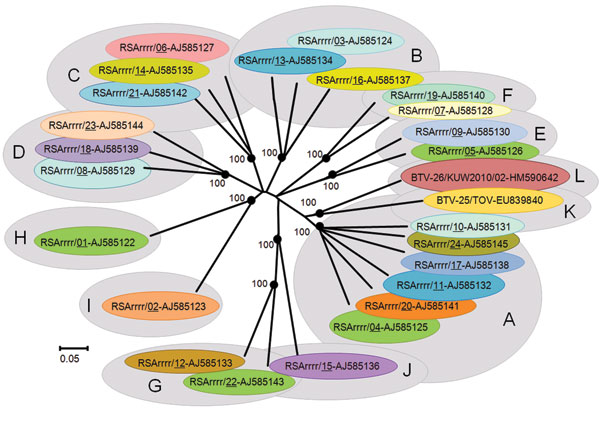
Figure 2. Neighbor-joining tree showing relationships between genome segment-2 (Seg-2) from KUW2010/02 with the 25 reference strains of different bluetongue virus (BTV) serotypes. The tree was constructed by using distance matrices, generated by using the p-distance determination algorithm in MEGA 4.1 (500 bootstrap replicates) (12). The 10 evolutionary branching points are indicated by black dots on the tree (along with their bootstrap values), which correlate with the 11 Seg-2 nucleotypes designated A–L. BTV-26 (KUW2010/02),forms a new 12th Seg-2 nucleotype (L). Members of the same Seg-2 nucleotype are characterized by 66.9% identity in their Seg-2 nucleotide sequences; members of different nucleotypes show 61.4% identity in Seg-2 (4). The scale bar indicates the number of substitutions per site. The tree based on the amino acid sequences of viral protein 2 showed very similar topology. GenBank accession numbers of Seg-2 used for comparative analyses: AJ585122–AJ585145, EU839840.
References
- Darpel KE, Batten CA, Veronesi E, Shaw AE, Anthony S, Bachanek-Bankowska K, Clinical signs and pathology shown by British sheep and cattle infected with bluetongue virus serotype 8 derived from the 2006 outbreak in northern Europe. Vet Rec. 2007;161:253–61. DOIPubMedGoogle Scholar
- Attoui H, Maan S, Anthony SJ, Mertens PPC. Bluetongue virus, other orbiviruses and other reoviruses: their relationships and taxonomy. In: Mellor PS, Baylis M, Mertens PPC, editors. Bluetongue monograph. 1st ed. London: Elsevier/Academic Press; 2009. p. 23–52.
- Mertens PPC, Pedley S, Cowley J, Burroughs JN, Corteyn AH, Jeggo MH, Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virology. 1989;170:561–5. DOIPubMedGoogle Scholar
- Maan S, Maan NS, van Rijn PA, van Gennip RGP, Sanders A, Wright IM, Full genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains. PLoS ONE. 2010;5:e10323. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. OIE 2009: final report of the 10th conference of the OIE Regional Commission for the Middle East Doha (Qatar), 26–29 October 2009 [cited 2011 Mar 11]. http://www.rr-middleeast.oie.int/download/pdf/Final%20Report_Doha_%20Qatar.pdf
- Shaw AE, Monaghan P, Alpar HO, Anthony S, Darpel KE, Batten CA, Development and validation of a real-time RT-PCR assay to detect genome bluetongue virus segment 1. J Virol Methods. 2007;145:115–26. DOIPubMedGoogle Scholar
- Toussaint JF, Sailleau C, Bréard E, Zientara S, De Clercq K. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J Virol Methods. 2007;140:115–23. DOIPubMedGoogle Scholar
- Orrù G, Ferrando ML, Meloni M, Liciardi M, Savini G, De Santis P. Rapid detection and quantitation of bluetongue virus (BTV) using a molecular beacon fluorescent probe assay. J Virol Methods. 2006;137:34–42. DOIPubMedGoogle Scholar
- Mertens PPC, Attoui H. Isolates of bluetongue virus (BTV) in the dsRNA virus collection at IAH Pirbright [updated 2010 Jun 23; cited 2010 Jul 25]. http://www.reoviridae.org/dsRNA_virus_proteins/ReoID/BTV-isolates.htm.
- Thevasagayam JA, Wellby MP, Mertens PP, Burroughs JN, Anderson J. Detection and differentiation of epizootic haemorrhagic disease of deer and bluetongue viruses by serogroup-specific sandwich ELISA. J Virol Methods. 1996;56:49–57. DOIPubMedGoogle Scholar
- Maan S, Rao S, Maan NS, Anthony SJ, Attoui H, Samuel AR, Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods. 2007a;143:132–9. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Grist NR, Ross CA, Bell EJ. Neutralization tests. In: Grist NR, Ross CA, Bell EJ, editors. Diagnostic methods in clinical virology, 2nd ed. London: Blackwell Scientific Publications; 1974. p. 64–79.
- Batten CA, Banyard A, King D, Henstock M, Edwards L, Sanders A, A real-time PCR assay for the specific detection of peste des petits ruminants virus. J Virol Methods. 2011;171:401–4. DOIPubMedGoogle Scholar
- Hofmann MA, Renzullo S, Mader M, Chaignat V, Worwa G, Thuer B. Genetic characterization of Toggenburg orbivirus, a new bluetongue virus from goats Switzerland. Emerg Infect Dis. 2008;14:1855–61. DOIPubMedGoogle Scholar