Volume 18, Number 12—December 2012
Dispatch
High Diversity of RNA Viruses in Rodents, Ethiopia
Figure 2
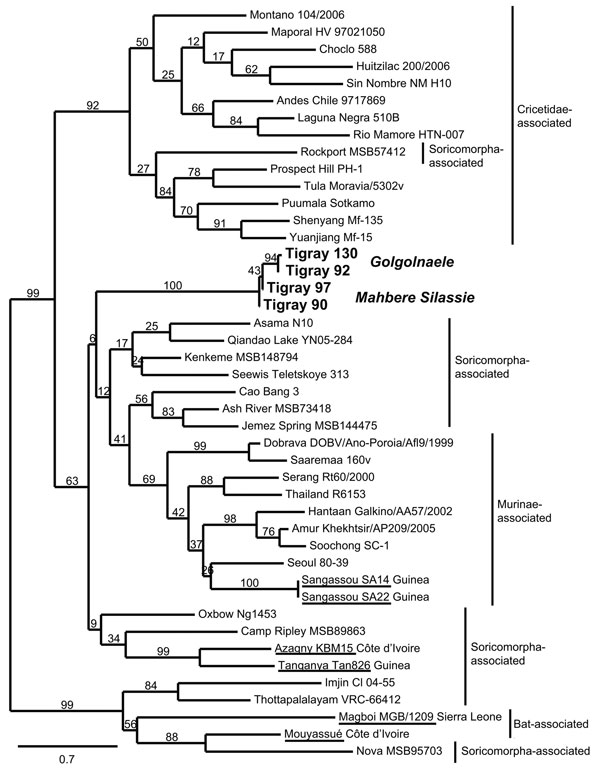
Figure 2. . Maximum-likelihood tree of hantaviruses showing the position of the 4 sequences of Tigray hantavirus (boldface; GenBank accession nos. JQ956484–JQ956487) found in kidney samples of Ethiopian white-footed mice (Stenocephalemys albipes). The tree was constructed on the basis of analysis of partial sequences of the RNA polymerase gene; phylogeny was estimated by using the maximum-likelihood method with the general time reversible + I + Γ (4 rate categories) substitution model to account for rate heterogeneity across sites as implemented in the PhyML program (8). Numbers represent percentage bootstrap support (1,000 replicates). Underlining indicates hantaviruses found in Africa. Scale bar indicates nucleotide substitutions per site. GenBank accession numbers of the virus strains: AB620030, NC_003468, EU929078, EF619961, JF276228, EF540771, EF543525, EF397003, NC_005235, AB620033, JN037851, FJ170809, FJ170812, AB620108, EF641807, FJ593501, GQ306150, AF005729, EU788002, AB620102, FJ593498, FJ593497, EF646763, NC_005225, GU566021, FJ809772, HM015221, AJ410618, DQ268652, JQ082305, EU424336, NC_005238, AM998806, NC_005217, DQ056292, EF050454, JN116261, EU001330, AJ005637, JQ287716.
References
- Woolhouse M, Gaunt E. Ecological origins of novel human pathogens. Crit Rev Microbiol. 2007;33:231–42. DOIPubMedGoogle Scholar
- Weiss S, Witkowski PT, Auste B, Nowak K, Weber N, Fahr J, Hantavirus in bat, Sierra Leone. Emerg Infect Dis. 2012;18:159–61. DOIPubMedGoogle Scholar
- Sumibcay L, Kadjo B, Gu SH, Kang HJ, Lim B, Cook J, Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Côte d’Ivoire. Virol J. 2012;9:34. DOIPubMedGoogle Scholar
- Gonzalez JP, McCormick JB, Baudon D, Gautun JP, Meunier DY, Dournon E, Serological evidence for Hantaan-related virus in Africa. Lancet. 1984;324:1036–7. DOIPubMedGoogle Scholar
- Klempa B, Koivogui L, Sylla O, Koulemou K, Auste B, Kruger DH, Serological evidence of human hantavirus infections in Guinea, West Africa. J Infect Dis. 2010;201:1031–4. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, Hantavirus in African wood mouse, Guinea. Emerg Infect Dis. 2006;12:838–40. DOIPubMedGoogle Scholar
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Goüy de Bellocq J, Borremans B, Katakweba A, Makundi R, Baird SJE, Becker-Ziaja B, Sympatric occurrence of 3 arenaviruses, Tanzania. Emerg Infect Dis. 2010;16:692–5. DOIPubMedGoogle Scholar
- Gonzalez JP, McCormick JB, Saluzzo JF, Herve JP, Georges AJ, Johnson KM. An arenavirus isolated from wild-caught rodents (Praomys species) in the Central African Republic. Intervirology. 1983;19:105–12. DOIPubMedGoogle Scholar
- Coulibaly-N’Golo D, Allali B, Kouassi SK, Fichet-Calvet E, Becker-Ziaja B, Rieger T, Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Côte d’Ivoire: implications for evolution of arenaviruses in Africa. PLoS ONE. 2011;6:e20893. DOIPubMedGoogle Scholar
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, Novel hantavirus sequences in shrew, Guinea. Emerg Infect Dis. 2007;13:520–2. DOIPubMedGoogle Scholar
- Kang HJ, Kadjo B, Dubey S, Jacquet F, Yanagihara R. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d’Ivoire. Virol J. 2011;8:373. DOIPubMedGoogle Scholar
- Coulaud X, Chouaib E, Georges AJ, Rollin P, Gonzalez JP. First human case of haemorrhagic fever with renal syndrome in the Central African Republic. Trans R Soc Trop Med Hyg. 1987;81:686. DOIPubMedGoogle Scholar
- Lecompte E, Aplin K, Denys C, Catzeflis F, Chades M, Chevret P. Phylogeny and biogeography of African Murinae based on mitochondrial and nuclear gene sequences, with a new tribal classification of the subfamily. BMC Evol Biol. 2008;8:199. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.