Volume 20, Number 12—December 2014
Dispatch
Francisella tularensis Bacteria Associated with Feline Tularemia in the United States
Figure
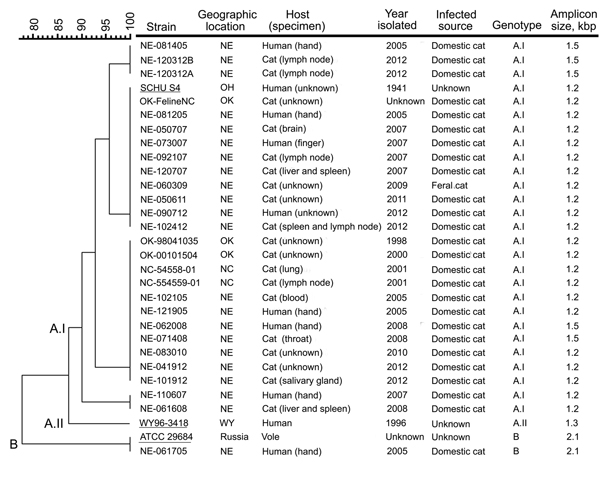
Figure. Molecular genotyping of Francisella tularensis isolates obtained from infected cats or humans bitten by an infected cat, United States, 1998–2012. Genotyping was performed by using pulsed-field gel electrophoresis (PFGE) and the PCR-based differential insertion sequence amplification (DISA) assay. A dendrogram of PFGE patterns obtained with PmeI-digested F. tularensis isolates is shown on the left; the scale bar at the top indicates distance in relative units. The genotype-specific amplicon lengths obtained with the DISA CR10 C+L+S primer set are shown on the right. Migration profiles of the PmeI restriction fragments from F. tularensis chromosomal DNA were normalized to SmaI-digested Staphylococcus aureus NCTC 8325 by using BioNumerics software (Applied Maths, Inc., Austin, TX, USA). Cluster analysis was performed by using the Dice correlation coefficient and the unweighted pair group mathematical average (UPGMA) clustering algorithm in the BioNumerics software. The DISA CR10 C+S primer pair identified the subtype A.I strains, whereas the CR10 C+L primer pair differentiated the subtype A.II and type B strains by the size of the amplicon produced. Underlining indicates F. tularensis strains SCHU S4 (subtype A.I), WY96–3418 (subtype A.II), and ATCC 29684 (type B), which were included as references. F. tularensis isolates NE-062508 and NE-073009 were no longer viable after storage and therefore not genotyped; these pathogens were isolated from domestic cats from eastern Nebraska (NE) in 2008 and 2009, respectively. ATCC, American Type Culture Collection; NC, North Carolina; OH, Ohio; OK, Oklahoma; WY, Wyoming.