Volume 20, Number 7—July 2014
Dispatch
New Viruses in Idiopathic Human Diarrhea Cases, the Netherlands
Figure 1
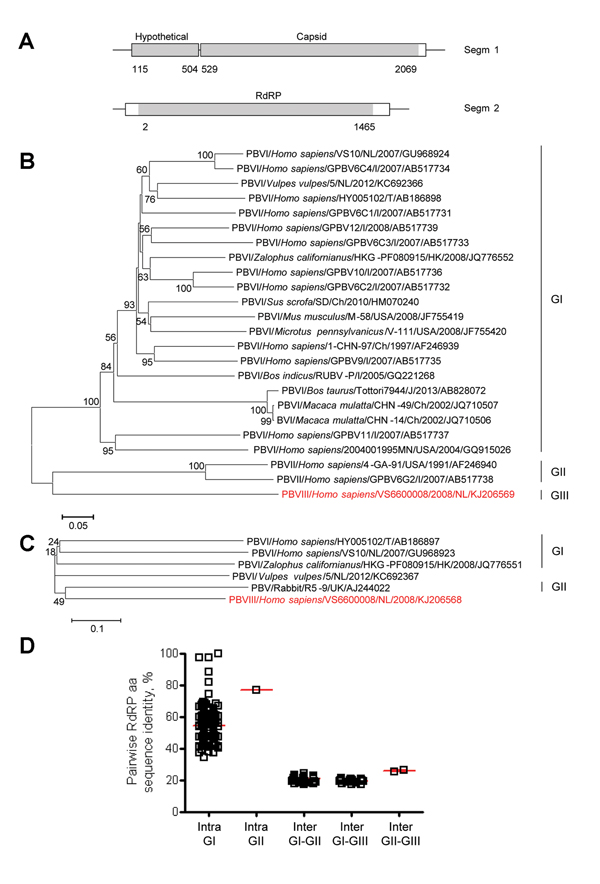
Figure 1. Genome organization and phylogenetic analysis of human picobirnavirus (PBV) VS6600008 isolated in the Netherlands, 2005–2009A) Putative schematic genome organization of human PBV VS6600008Locations of major open reading frames are indicated in white and sequences obtained by next-generation sequencing are indicated in graySegm, segment; RdRP, RNA-dependent RNA polymeraseB) Phylogenetic neighbor-joining tree with p-distances and 1,000 bootstrap replicates of amino acid sequences of partial RdRP genes corresponding to aa 80–377 of reference PBV strain HY005102; AB186898, PBV VS6600008, and representative PBVsAlignments were created by using ClustalX 2.0 (http://www.clustal.org/)Viruses are shown as virus/host species/strain/country/year/GenBank accession no(if available)Virus isolated in this study is indicated in redGenogroups are indicated on the rightScale bar indicates amino acid substitutions per siteNL, the Netherlands; I, India; T, Thailand; Ch, China; USA, United States; UK, United Kingdom; J, Japan; HK, Hong KongC) Phylogenetic neighbor-joining tree with p-distances and 1,000 bootstrap replicates of the amino acid sequences of the partial capsid genes corresponding to aa 1–220 of reference PBV strain HY005102; AB186897, PBV VS6600008, and representative PBVsAlignments were created by using ClustalX 2.0Virus isolated in this study is indicated in redGenogroups are indicated on the rightScale bar indicates amino acid substitutions per siteD) Pairwise intragenogroup (Intra) and intergenogroup (Inter) amino acid sequence identities determined by using Bioedit 7.0.9.0 (http://www.mbio.ncsu.edu/bioedit/bioedit.html) between the partial RdRP sequences (corresponding to amino acids 80–377 of reference PBV strain HY005102; AB186898)Each square represents pairwise RdRP amino acid sequence identity between viruses in panel BRed bars indicate mean and SEM.