Rapid Whole-Genome Sequencing for Surveillance of Salmonella enterica Serovar Enteritidis
Henk C. den Bakker, Marc W. Allard, Dianna Bopp, Eric W. Brown, John Fontana, Zamin Iqbal, Aristea Kinney, Ronald Limberger, Kimberlee A. Musser, Matthew Shudt, Errol Strain, Martin Wiedmann, and William J. Wolfgang

Author affiliations: Cornell University, Ithaca, New York, USA (H. den Bakker, M. Wiedmann); US Food and Drug Administration, College Park, Maryland, USA (M.W. Allard, E.W. Brown, E. Strain); New York State Department of Health/Wadsworth Center, Albany, New York, USA (D. Bopp, R. Limberger, M. Shudt, K.A. Musser, W.J. Wolfgang); Connecticut Department of Public Health, Rocky Hill, Connecticut, USA (J. Fontana, A. Kinney); Wellcome Trust Centre for Human Genetics, Oxford, UK (Z. Iqbal)
Main Article
Figure 1
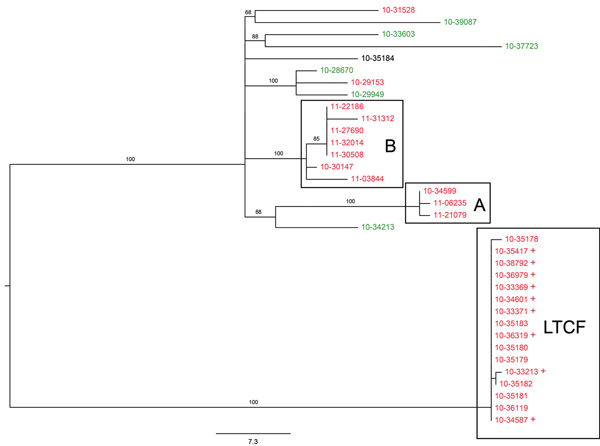
Figure 1.
Figure 1. Maximum-likelihood tree of population structure of Salmonella enterica serovar Enteritidis isolates obtained in New York and neighboring states, USA. The tree was inferred by using a general time-reversible model with a gamma distribution and was inferred to be the best fit model by the maximum-likelihood method implemented in MEGA 5.1 (22). Values on branches are bootstrap values based on 150 bootstrap replicates. Note the well supported and distant cluster associated with the long-term care facility (LTCF), as well as additional clusters A and B. Labels of isolates are colored according to their New York State Department of Health Wadsworth Laboratories multilocus variable-number tandem-repeat analysis (MLVA) subtype designation. Green, MLVA subtype B; red, MLVA subtype W; black, MLVA subtype AE. + indicates isolates in the LTCF cluster that were detected only by whole-genome analysis and were not detected epidemiologically. Scale bar indicates single-nucleotide polymorphisms per site.
Main Article
Page created: July 18, 2014
Page updated: July 18, 2014
Page reviewed: July 18, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
