Volume 20, Number 8—August 2014
Dispatch
Rapid Detection, Complete Genome Sequencing, and Phylogenetic Analysis of Porcine Deltacoronavirus
Figure
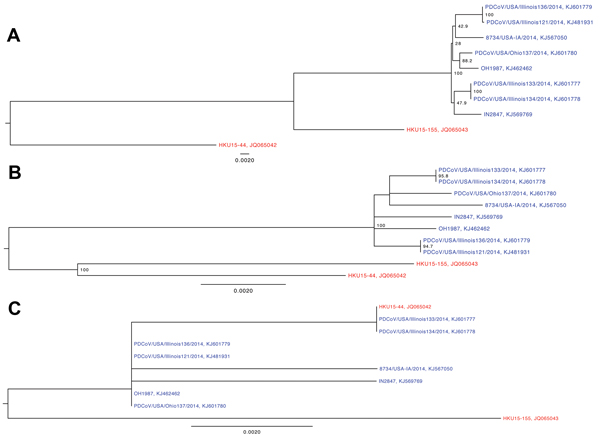
Figure. Phylogenetic trees of the complete porcine deltacoronavirus (PDCoV) genome (A), spike gene (B), and nonstructural protein 6 (NS6) accessory gene (C)US strains are in blue; China strains are in redBootstrap values >70% are illustratedScale bar indicates nucleotide substitutions per site.
Page created: April 29, 2014
Page updated: April 29, 2014
Page reviewed: April 29, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.