Genomic Epidemiology of Salmonella enterica Serotype Enteritidis based on Population Structure of Prevalent Lineages
Xiangyu Deng

, Prerak T. Desai, Henk C. den Bakker, Matthew Mikoleit, Beth Tolar, Eija Trees, Rene S. Hendriksen, Jonathan G. Frye, Steffen Porwollik, Bart C. Weimer, Martin Wiedmann, George M. Weinstock, Patricia I. Fields
1, and Michael McClelland
1
Author affiliations: University of Georgia, Griffin, Georgia, USA (X. Deng); University of California Irvine, Irvine, California, USA (P.T. Desai, S. Porwollik, M. McClelland); Cornell University, Ithaca, New York, USA (H.C. den Bakker, M. Wiedmann); Centers for Disease Control and Prevention, Atlanta, Georgia, USA (M. Mikoleit, B. Tolar, E. Trees, P.I. Fields); Technical University of Denmark, Lyngby, Denmark (R.S. Hendriksen); US Department of Agriculture, Athens, Georgia, USA (J.G. Frye); University of California Davis, Davis, California, USA (B.C. Weimer); Washington University School of Medicine, St. Louis, Missouri, USA (G.M. Weinstock)
Main Article
Figure 4
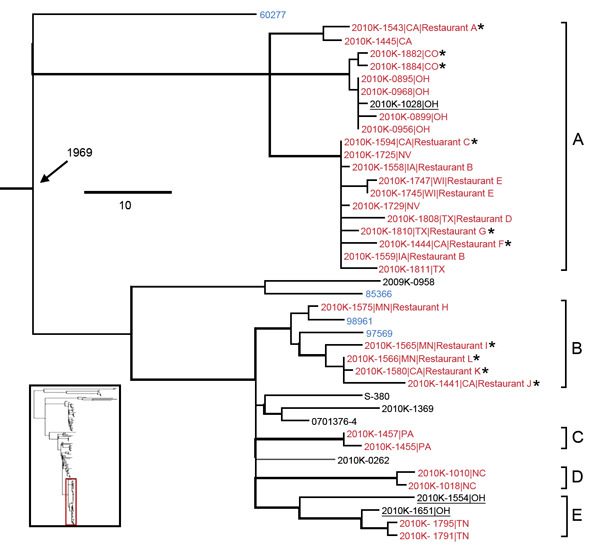
Figure 4. Salmonella enterica serotype Enteritidis clades associated with the 2010 US shelled eggs outbreak. Red indicates isolates sequenced as part of the outbreak investigation using Roche 454 technology (Indianapolis, IN, USA); blue indicates isolates associated with chicken or chicken products; asterisk (*) indicates the isolate was traced back to the implicated egg production facility. Five outbreak clades are highlighted and designated as A–E, of which A and B are definite and C, D, and E are putative. Isolates ascribed to the egg outbreak in this study are underlined. Branches with bootstrap values >0.9 are shown by thickened lines. Age of the ancestral node (median most recent common ancestor) is labeled. Scale bar indicates single-nucleotide polymorphisms. Inset indicates the location of the highlighted lineage on the global phylogenetic tree.
Main Article
Page created: August 13, 2014
Page updated: August 13, 2014
Page reviewed: August 13, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
