Volume 21, Number 12—December 2015
Dispatch
Japanese Macaques (Macaca fuscata) as Natural Reservoir of Bartonella quintana
Figure 2
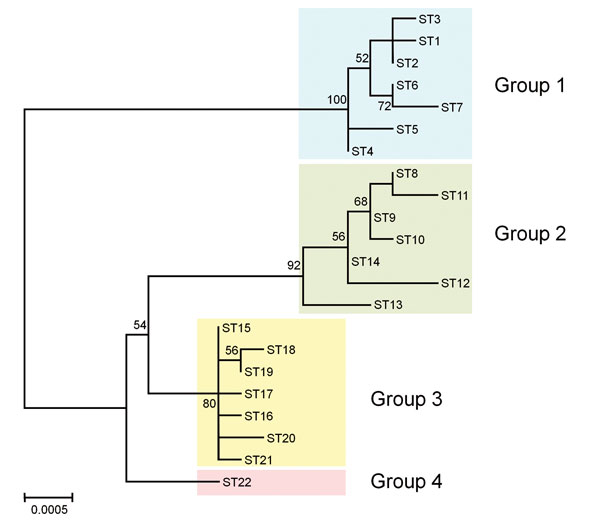
Figure 2. Phylogenetic tree showing the genetic relationship among Bartonella quintana strains from humans and macaques. The tree was constructed from the concatenated sequences (4,270 bp) of the 9 loci used for multilocus sequence typing by using the maximum-likelihood method based on the Tamura 3-parameter model in MEGA6 (13). The 22 sequence types (STs) of B. quintana strains from humans (STs 1–7), cynomolgus macaques (STs 8–4), rhesus macaques (STs 15–21), and Japanese macaques (ST22) were included in the tree. Colored rectangles show 4 groups classified by host species. The scale bar indicates estimated evolutionary distance. Bootstrap values were obtained with 1,000 replicates. Only bootstrap replicates >50% are noted.
References
- Foucault C, Brouqui P, Raoult D. Bartonella quintana characteristics and clinical management. Emerg Infect Dis. 2006;12:217–23 . DOIPubMedGoogle Scholar
- Deng H, Le Rhun D, Buffet JP, Cotte V, Read A, Birtles RJ, Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet Res. 2012;43:15. DOIPubMedGoogle Scholar
- O’Rourke LG, Pitulle C, Hegarty BC, Kraycirik S, Killary KA, Grosenstein P, Bartonella quintana in cynomolgus monkey (Macaca fascicularis). Emerg Infect Dis. 2005;11:1931–4. DOIPubMedGoogle Scholar
- Maggi RG, Mascarelli PE, Balakrishnan N, Rohde CM, Kelly CM, Ramaiah L, “Candidatus Mycoplasma haemomacaque” and Bartonella quintana bacteremia in cynomolgus monkeys. J Clin Microbiol. 2013;51:1408–11. DOIPubMedGoogle Scholar
- Huang R, Liu Q, Li G, Li D, Song X, Birtles RJ, Bartonella quintana infections in captive monkeys, China. Emerg Infect Dis. 2011;17:1707–9. DOIPubMedGoogle Scholar
- Li H, Bai JY, Wang LY, Zeng L, Shi YS, Qiu ZL, Genetic diversity of Bartonella quintana in macaques suggests zoonotic origin of trench fever. Mol Ecol. 2013;22:2118–27. DOIPubMedGoogle Scholar
- Fooden J, Aimi M. Systematic review of Japanese macaques, Macaca fuscata (Gray, 1870). Fieldiana Zoology. 2005;104:1–198.
- Zhang P, Chomel BB, Schau MK, Goo JS, Droz S, Kelminson KL, A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc Natl Acad Sci U S A. 2004;101:13630–5 . DOIPubMedGoogle Scholar
- Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803 .PubMedGoogle Scholar
- Kabeya H, Inoue K, Izumi Y, Morita T, Imai S, Maruyama S. Bartonella species in wild rodents and fleas from them in Japan. J Vet Med Sci. 2011;73:1561–7. DOIPubMedGoogle Scholar
- Roux V, Raoult D. The 16S–23S rRNA intergenic spacer region of Bartonella (Rochalimaea) species is longer than usually described in other bacteria. Gene. 1995;156:107–11. DOIPubMedGoogle Scholar
- Arvand M, Raoult D, Feil EJ. Multi-locus sequence typing of a geographically and temporally diverse sample of the highly clonal human pathogen Bartonella quintana. PLoS ONE. 2010;5:e9765. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res. 2009;40:29. DOIPubMedGoogle Scholar
- Bittar F, Keita MB, Lagier JC, Peeters M, Delaporte E, Raoult D. Gorilla gorilla gorilla gut: a potential reservoir of pathogenic bacteria as revealed using culturomics and molecular tools. Sci Rep. 2014;4:7174.
Page created: November 17, 2015
Page updated: November 17, 2015
Page reviewed: November 17, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.