Volume 21, Number 2—February 2015
Dispatch
Comparative Analysis of African Swine Fever Virus Genotypes and Serogroups
Figure 1
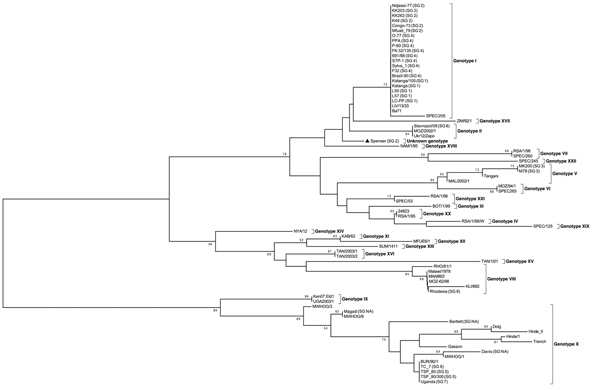
Figure 1. Phylogenetic tree of African swine fever virus (ASFV) isolates maintained in a collection at the National Research Institute for Veterinary Virology and Microbiology in Рokrov, Russia; the variable part of B646L gene relative to the 22 known p72 genotypes (labeled I-XXII) was used for analysis. The tree was reconstructed by using the minimum evolution method with 1,000 replicates.
1These authors contributed equally to this article.
Page created: January 21, 2015
Page updated: January 21, 2015
Page reviewed: January 21, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.