Volume 21, Number 2—February 2015
Research
Microbiota That Affect Risk for Shigellosis in Children in Low-Income Countries
Figure 3
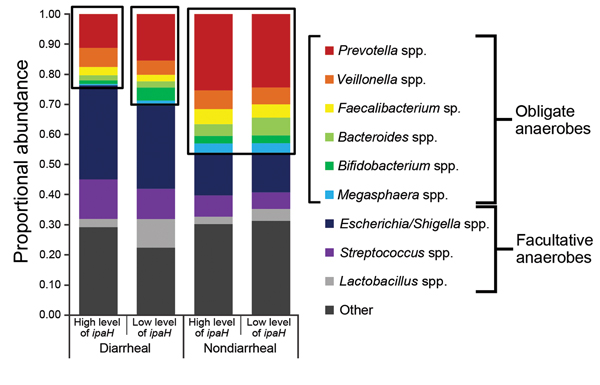
Figure 3. Overall 16S rRNA gene–based bacterial community profiles (proportional abundance) of diarrheal samples with high levels of ipaH gene (n = 277), diarrheal samples with low levels of ipaH gene (n = 1,023), nondiarrheal samples with high levels of ipaH gene (n = 127), and nondiarrheal samples with low levels of ipaH gene (n = 1,608) from children in low-income countries. Other indicates sequences that were not identified as 1 of the 9 most abundant taxa or did not have good (>100 bp exact match, >97% identity) matches with isolate sequences from the Ribosomal Database Project (19).
References
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. DOIPubMedGoogle Scholar
- Walker CL, Aryee MJ, Boschi-Pinto C, Black RE. Estimating diarrhea mortality among young children in low and middle income countries. PLoS ONE. 2012;7:e29151. DOIPubMedGoogle Scholar
- Lindsay B, Ramamurthy T, Sen Gupta S, Takeda Y, Rajendran K, Nair GB, Diarrheagenic pathogens in polymicrobial infections. Emerg Infect Dis. 2011;17:606–11. DOIPubMedGoogle Scholar
- Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, Yang Z, Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis. 2013;208:1794–802. DOIPubMedGoogle Scholar
- Platts-Mills JA, Liu J, Houpt ER. New concepts in diagnostics for infectious diarrhea. Mucosal Immunol. 2013;6:876–85. DOIPubMedGoogle Scholar
- Pop M, Walker AW, Paulson JN, Lindsay B, Antonio M, Hossain MA, Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiome composition. Genome Biol. 2014;15:R76. DOIPubMedGoogle Scholar
- Lindsay B, Ochieng JB, Ikumapayi UN, Toure A, Ahmed D, Li S, Quantitative PCR for detection of Shigella improves ascertainment of Shigella burden in children with moderate-to-severe diarrhea in low-income countries. J Clin Microbiol. 2013;51:1740–6. DOIPubMedGoogle Scholar
- Platts-Mills JA, Gratz J, Mduma E, Svensen E, Amour C, Liu J, Association between stool enteropathogen quantity and disease in Tanzanian children using TaqMan array cards: a nested case-control study. Am J Trop Med Hyg. 2014;90:133–8. DOIPubMedGoogle Scholar
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22. DOIPubMedGoogle Scholar
- Bhavnani D, Goldstick JE, Cevallos W, Trueba G, Eisenberg JNS. Synergistic effects between rotavirus and coinfecting pathogens on diarrheal disease: evidence from a community-based study in northwestern Ecuador. Am J Epidemiol. 2012;176:387–95. DOIPubMedGoogle Scholar
- Yun J-H, Yim D-S, Kang J-Y, Kang B-Y, Shin E-A, Chung M-J, Identification of Lactobacillus ruminus SPM0211 isolated from healthy Koreans and its antimicrobial activity against some pathogens. Arch Pharm Res. 2005;28:660–6. DOIPubMedGoogle Scholar
- Guandalini S. Probiotics for children with diarrhea: an update. J Clin Gastroenterol. 2008;42(Suppl 2):S53–7. DOIPubMedGoogle Scholar
- Srikanth CV, McCormick BA. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: a three-way crosswalk. Interdiscip Perspect Infect Dis. 2008;2008:626827. . Epub 2008 Oct 29.DOIGoogle Scholar
- Levine MM, Kotloff KL, Nataro JP, Muhsen K. The Global Enteric Multicenter Study (GEMS): impetus, rationale, and genesis. Clin Infect Dis. 2012;55(Suppl 4):S215–24. DOIPubMedGoogle Scholar
- Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH, van Eijk A, The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis. 2012;55(Suppl 4):S232–45. DOIPubMedGoogle Scholar
- Panchalingam S, Antonio M, Hossain A, Mandomando I, Ochieng B, Oundo J, Diagnostic microbiologic methods in the GEMS-1 case/control study. Clin Infect Dis. 2012;55(Suppl 4):S294–302. DOIPubMedGoogle Scholar
- Vu DT, Sethabutr O, Von Seidlein L, Tran VT, Do GC, Bui TC, Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J Clin Microbiol. 2004;42:2031–5. DOIPubMedGoogle Scholar
- Ghodsi M, Liu B, Pop M. DNACLUST: accurate and efficient clustering of phylogenetic marker genes. BMC Bioinformatics. 2011;12:271 and. DOIPubMedGoogle Scholar
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–6. DOIPubMedGoogle Scholar
- Rothman KJ. Epidemiology: an introduction. New York: Oxford University Press; 2012.
- Knol MJ, VanderWeele TJ. Recommendations for presenting analyses of effect modification and interaction. Int J Epidemiol. 2012;41:514–20. DOIPubMedGoogle Scholar
- Andersson T, Alfredsson L, Källberg H, Zdravkovic S, Ahlbom A. Calculating measures of biological interaction. Eur J Epidemiol. 2005;20:575–9. DOIPubMedGoogle Scholar
- Zou GY. On the estimation of additive interaction by use of the four-by-two table and beyond. Am J Epidemiol. 2008;168:212–24. DOIPubMedGoogle Scholar
- Hosmer DW, Lemeshow S. Confidence interval estimation of interaction. Epidemiology. 1992;3:452–6 and. DOIPubMedGoogle Scholar
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Vegan: Community Ecology Package, 2013 [cited 2014 Apr 8]. http://cran.r-project.org/web/packages/vegan/index.html
- Lindsay B, Pop M, Antonio M, Walker AW, Mai V, Ahmed D, Survey of culture, goldengate assay, universal biosensor assay, and 16S rRNA gene sequencing as alternative methods of bacterial pathogen detection. J Clin Microbiol. 2013;51:3263–9. DOIPubMedGoogle Scholar
- Sinha A, SenGupta S, Guin S, Dutta S, Ghosh S, Mukherjee P, Culture-independent real-time PCR reveals extensive polymicrobial infections in hospitalized diarrhoea cases in Kolkata, India. Clin Microbiol Infect. 2013;19:173–80. DOIPubMedGoogle Scholar
- Kwambana BA, Ikumapayi UN, Sallah N, Dione M, Jarju S, Panchalingham S, High genotypic diversity among rotavirus strains infecting Gambian children. Pediatr Infect Dis J. 2014;33(Suppl 1):S69–75. DOIPubMedGoogle Scholar
- Lynn HS, McCulloch CE. When does it pay to break the matches for analysis of a matched-pairs design? Biometrics. 1992;48:397–409. DOIPubMedGoogle Scholar
- Hansson L, Khamis HJ. Matched samples logistic regression in case-control studies with missing values: when to break the matches. Stat Methods Med Res. 2008;17:595–607 . DOIPubMedGoogle Scholar
Page created: January 20, 2015
Page updated: January 20, 2015
Page reviewed: January 20, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.