Reassortant Highly Pathogenic Influenza A(H5N6) Virus in Laos
Frank Y.K. Wong

, Phouvong Phommachanh, Wantanee Kalpravidh, Chintana Chanthavisouk, Jeffrey Gilbert, John Bingham, Kelly R. Davies, Julie Cooke, Debbie Eagles, Sithong Phiphakhavong, Songhua Shan, Vittoria Stevens, David T. Williams, Phachone Bounma, Bounkhouang Khambounheuang, Christopher Morrissy, Bounlom Douangngeun, and Subhash Morzaria
Author affiliations: Commonwealth Scientific and Industrial Research Organisation Australian Animal Health Laboratory, Geelong, Victoria, Australia (F.Y.K. Wong, J. Bingham, K.R. Davies, J. Cooke, D. Eagles, S. Shan, V. Stevens, D.T. Williams, C. Morrissy); National Animal Health Laboratory, Vientiane, Laos (P. Phommachanh, B. Douangngeun); Ministry of Agriculture and Forestry, Vientiane (S. Phiphakhavong, P. Bounma, B. Khambounheuang); Food and Agricultural Organization of the United Nations Regional Office for Asia and the Pacific, Bangkok, Thailand (W. Kalpravidh, C. Chanthavisouk, J. Gilbert, S. Morzaria)
Main Article
Figure 2
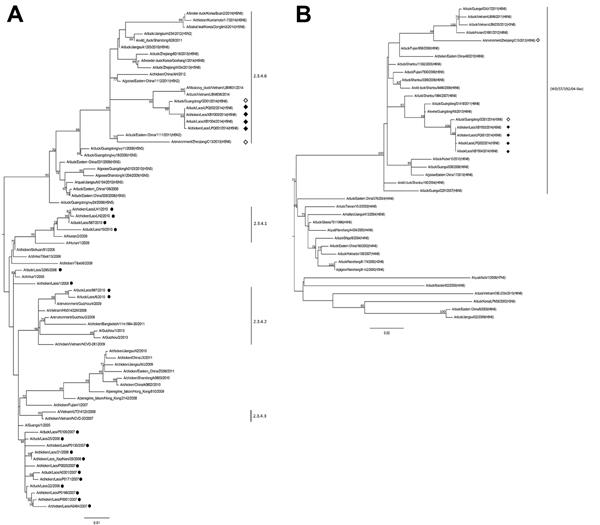
Figure 2. Phylogenetic analyses of influenza A(H5N6) viruses detected in Laos, March 2014, on the basis of the hemagglutinin (HA) and N6 neuraminidase (NA) genes. A) HA subtree showing relationships of emergent influenza A(H5N6) viruses with clade 2.3.4 H5 avian influenza viruses and B) NA subtree showing relationships with Asian lineage N6 avian influenza viruses. Vertical lines denote H5 subtype virus clades on the HA tree and the WD/ST/192/04 (A/wild duck/Shantou/192/2004)-like N6 gene pool on the NA tree. Proposed clade 2.3.4.6 has not been formally recognized by the World Health Organization/World Organisation for Animal Health/Food and Agricultural Organization of the United Nations H5N1 Evolution Working Group. Black diamonds indicate viruses identified in this study, white diamonds indicate Asian influenza A(H5N6) viruses identified in other studies, and black circles indicate viruses previously identified in Laos. All viruses are subtype H5N1 unless otherwise indicated. Bootstrap values ≥70% from 1,000 replicates are indicated at relevant nodes, and scale bars indicate nucleotide substitutions per site. The full HA and NA trees are provided in the online Technical Appendix Figure, panels A and B (http://wwwnc.cdc.gov/EID/article/21/3/14-1488-Techapp1.pdf).
Main Article
Page created: February 18, 2015
Page updated: February 18, 2015
Page reviewed: February 18, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
