Mycobacterium bovis in Panama, 2013
Fermín Acosta, Ekatherina Chernyaeva, Libardo Mendoza, Dilcia Sambrano, Ricardo Correa, Mikhail Rotkevich, Miroslava Tarté, Humberto Hernández, Bredio Velazco, Cecilia de Escobar, Jacobus H. de Waard, and Amador Goodridge

Author affiliations: Instituto de Investigaciones Científicas y Servicios de Alta Tecnología, Ciudad del Saber, Panama (F. Acosta, D. Sambrano, R. Correa, A. Goodridge); Petersburg Institute of Phthisiopulmonology, St. Petersburg, Russia (E. Chernyaeva); St. Petersburg State University, St. Petersburg (E. Chernyaeva, M. Rotkevich); Inversiones para el Desarrollo de Coclé S.A., Anton, Panama (L. Mendoza); Ministerio de Desarrollo Agropecuario, Tocumen, Panama (M. Tarté, H. Hernández, B. Velazco); Instituto de Investigaciones Agropecuarias de Panamá, Ciudad del Saber (C. de Escobar); Instituto de Biomedicina, Universidad Central de Venezuela, Caracas, Venezuela. (J.H. de Waard)
Main Article
Figure 2
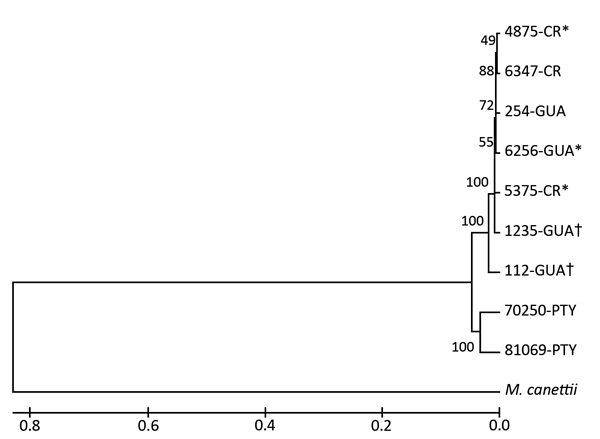
Figure 2.
Phylogenic placement of the Mycobacterium bovis isolates. The M. bovis AF2122/97 sequence was used as a reference for single-nucleotide polymorphism analysis (GenBank accession no. NC_002945.3).The tree was derived from an unweighted pair-grouping method analysis algorithm by using DNA fragment sequence analysis. Numbers on branches represent bootstrap percentages from 500 replicates. M. canettii was used as the outgroup. Evolutionary analyses were performed by using MEGA6 software (http://www.megasoftware.net). Symbols indicate M. bovis isolates with identical genotypes according to mycobacterial interspersed repetitive units–variable number of tandem repeats. Scale bar indicates mean distances between strains according to base substitutions (%). CR, Costa Rica; GUA, Guatemala, PTY, Coclé, Panama.
Main Article
Page created: April 29, 2015
Page updated: April 29, 2015
Page reviewed: April 29, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
