Volume 21, Number 7—July 2015
Dispatch
Swine Influenza A(H3N2) Virus Infection in Immunocompromised Man, Italy, 2014
Figure 2
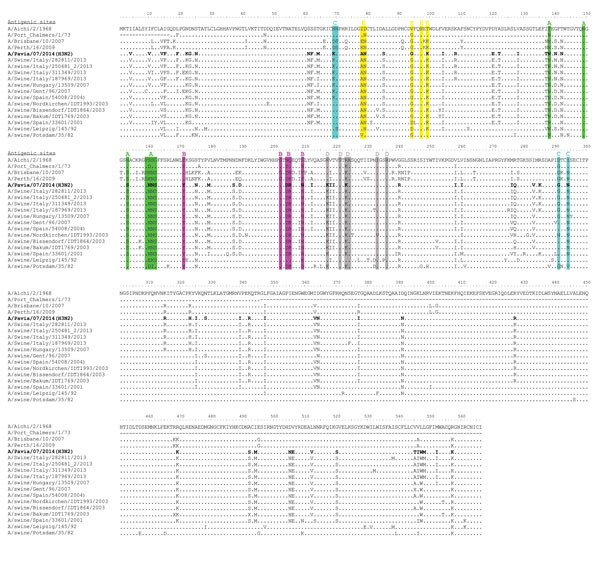
Figure 2. Amino acid sequence alignment of the hemagglutinin protein of swine influenza virus (SIV) (H3N2) strain A/Pavia/07/2014 (bold) and other SIVs. Antigenic sites A, B, C, D, and E of H3 HA are highlighted in green, magenta, blue, gray, and yellow, respectively, as proposed by others (11). Amino acid changes with respect to the A/Aichi/2/1968 strain are indicated for each strain.
References
- Zell R, Scholtissek C, Ludwig S. Genetics, evolution, and the zoonotic capacity of European swine influenza viruses. Curr Top Microbiol Immunol. 2013;370:29–55. DOIPubMedGoogle Scholar
- Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis. 2007;44:1084–8. DOIPubMedGoogle Scholar
- Krumbholz A, Lange J, Dürrwald R, Walther M, Muller TH, Kuhnel D, Prevalence of antibodies to European porcine influenza viruses in humans living in high pig density areas of Germany. Med Microbiol Immunol (Berl). 2014;203:13–24. DOIPubMedGoogle Scholar
- Gerloff NA, Kremer JR, Charpentier E, Sausy A, Olinger CM, Weicherding P, Swine influenza virus antibodies in humans, western Europe, 2009. Emerg Infect Dis. 2011;17:403–11. DOIPubMedGoogle Scholar
- Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of human respiratory picornavirus species, including the newly identified group C rhinoviruses, during a 1-year surveillance of a hospitalized patient population in Italy. J Clin Microbiol. 2011;49:373–6. DOIPubMedGoogle Scholar
- World Health Organization. CDC protocol of realtime RTPCR for influenza A(H1N1). 2009 Oct 6 [cited 2009 Dec 15]. http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. DOIPubMedGoogle Scholar
- Lycett SJ, Baillie G, Coulter E, Bhatt S, Kellam P, McCauley JW, Estimating reassortment rates in co-circulating Eurasian swine influenza viruses. J Gen Virol. 2012;93:2326–36. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Tharakaraman K, Raman R, Stebbins NW, Viswanathan K, Sasisekharan V, Sasisekharan R. Antigenically intact hemagglutinin in circulating avian and swine influenza viruses and potential for H3N2 pandemic. Sci Rep. 2013;3:1822.
- World Organisation for Animal Health. Manual of diagnostic tests and vaccines for terrestrial animals. Chapter 2.08.08. Swine influenza [cited 2014 Jun 6]. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.08.08_SWINE_INFLUENZA.pdf
- Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Genetic reassortant between avian and human influenza a viruses in Italian pigs. Virology. 1993;193:503–6. DOIPubMedGoogle Scholar
- Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis. 2013;57(Suppl 1):S4–11. DOIPubMedGoogle Scholar
- Piralla A, Gozalo-Margüello M, Fiorina L, Rovida F, Muzzi A, Colombo AA, Different drug-resistant influenza A(H3N2) variants in two immunocompromised patients treated with oseltamivir during the 2011–2012 influenza season in Italy. J Clin Virol. 2013;58:132–7. DOIPubMedGoogle Scholar
1Members of the Influenza Surveillance Study Group who contributed data are listed at the end of this article.
Page created: June 15, 2015
Page updated: June 15, 2015
Page reviewed: June 15, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.