Volume 22, Number 10—October 2016
Dispatch
Chikungunya Virus in Febrile Humans and Aedes aegypti Mosquitoes, Yucatan, Mexico
Figure
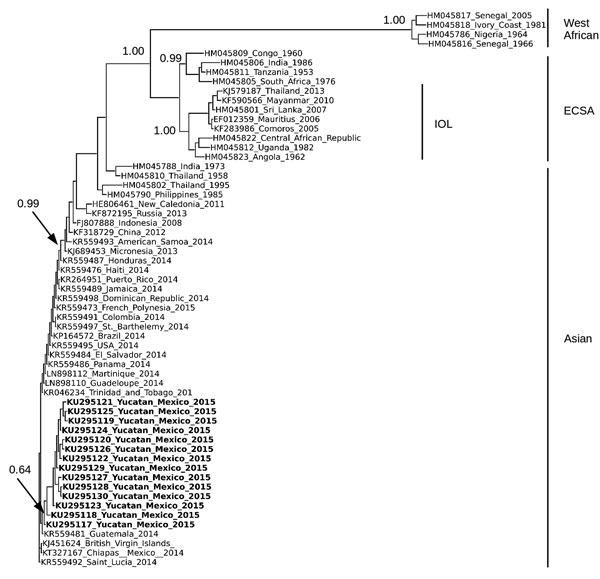
Figure. Phylogenetic analysis of chikungunya virus (CHIKV) isolates from Yucatan, Mexico. Analysis was based on a 3,744-nt structural gene region (capsid-E3-E2-6K-E1) of 63 CHIKV isolates, including the 14 isolates from Yucatan. Sequences were aligned by using MUSCLE (11), and the tree was constructed by using the neighbor-joining algorithm as implemented in PHYLIP (12) and using ETE3 (Environment for Tree Exploration 3) (13). Isolates are identified by GenBank accession number, country, and year isolated. CHIKV isolates from the Yucatan are shown in bold. Bootstrap values were generated by using 1,000 repetitions and normalized on a scale of 0–1. Bootstrap values for select branches are shown. 6K, membrane-associated peptide; E, envelope; ECSA, East/Central/South African lineage; IOL, Indian Ocean lineage.
References
- Weaver SC, Forrester NL. Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res. 2015;120:32–9. DOIPubMedGoogle Scholar
- Powers AM. Risks to the Americas associated with the continued expansion of chikungunya virus. J Gen Virol. 2015;96:1–5. DOIPubMedGoogle Scholar
- Rivera-Ávila RC. Chikungunya fever in Mexico: confirmed case and notes on the epidemiologic response [in Spanish]. Salud Publica Mex. 2014;56:402–4.PubMedGoogle Scholar
- Díaz-Quinonez JA, Ortiz-Alcantara J, Fragoso-Fonseca DE, Garces-Ayala F, Escobar-Escamilla N, Vazquez-Pichardo M, Complete genome sequences of chikungunya virus strains isolated in Mexico: first detection of imported and autochthonous cases. Genome Announc. 2015;3:e00300–15. DOIPubMedGoogle Scholar
- Kautz TF, Diaz-Gonzalez EE, Erasmus JH, Malo-Garcia IR, Langsjoen RM, Patterson EI, Chikungunya virus as cause of febrile illness outbreak, Chiapas, Mexico, 2014. Emerg Infect Dis. 2015;21:2070–3. DOIPubMedGoogle Scholar
- Díaz-González EE, Kautz TF, Dorantes-Delgado A, Malo-Garcia IR, Laguna-Aguilar M, Langsjoen RM, First report of Aedes aegypti transmission of chikungunya virus in the Americas. Am J Trop Med Hyg. 2015;93:1325–9. DOIPubMedGoogle Scholar
- Darsie RF Jr. A survey and bibliography of the mosquito fauna of Mexico (Diptera: Culicidae). J Am Mosq Control Assoc. 1996;12:298–306.PubMedGoogle Scholar
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–51.PubMedGoogle Scholar
- Tsetsarkin KA, Weaver SC. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7:e1002412. DOIPubMedGoogle Scholar
- Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. DOIPubMedGoogle Scholar
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. DOIPubMedGoogle Scholar
- Felsenstein J. PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics. 1989;5:164–6.
- Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. DOIPubMedGoogle Scholar
- Chang S-F, Su C-L, Shu P-Y, Yang C-F, Liao T-L, Cheng C-H, Concurrent isolation of chikungunya virus and dengue virus from a patient with coinfection resulting from a trip to Singapore. J Clin Microbiol. 2010;48:4586–9. DOIPubMedGoogle Scholar
- Parreira R, Centeno-Lima S, Lopes A, Portugal-Calisto D, Constantino A, Nina J. Dengue virus serotype 4 and chikungunya virus coinfection in a traveller returning from Luanda, Angola, January 2014. Euro Surveill. 2014;19:20730. DOIPubMedGoogle Scholar