Volume 22, Number 11—November 2016
Research
Ambulatory Pediatric Surveillance of Hand, Foot and Mouth Disease as Signal of an Outbreak of Coxsackievirus A6 Infections, France, 2014–2015
Abstract
The clinical impact of enteroviruses associated with hand, foot and mouth disease (HFMD) is unknown outside Asia, and the prevalence of enterovirus A71 (EV-A71) in particular might be underestimated. To investigate the prevalence of enterovirus serotypes and the clinical presentations associated with HFMD in France, we conducted prospective ambulatory clinic–based surveillance of children during April 2014–March 2015. Throat or buccal swabs were collected from children with HFMD and tested for the enterovirus genome. Physical examinations were recorded on a standardized form. An enterovirus infection was detected in 523 (79.3%) of 659 children tested. Two epidemic waves occurred, dominated by coxsackievirus (CV) A6, which was detected in 53.9% of enterovirus-infected children. CV-A6 was more frequently related to atypical HFMD manifestations (eruptions extended to limbs and face). Early awareness and documentation of HFMD outbreaks can be achieved by syndromic surveillance of HFMD by ambulatory pediatricians and rapid enterovirus testing and genotyping.
Although mostly asymptomatic or self-limited, enterovirus infections comprise a wide spectrum of clinical manifestations in children, which can require medical attention. Periodically, the emergence of an enterovirus serotype is associated with outbreaks of more serious disease resulting in serious illness and even death. Recent examples are the emergence of enterovirus A71 (EV-A71), which was responsible for large hand, foot and mouth disease (HFMD) outbreaks associated with rare but severe rhombencephalitis in Asia, and an EV-D68 epidemic associated with severe respiratory infections (1,2). Monitoring enterovirus infections and providing laboratory confirmation of the serotypes associated with different clinical presentations are of value for the early detection and awareness of emerging enterovirus infections (3,4).
EV-A71 is considered to be the most important neurotropic enterovirus in Southeast Asia countries, and EV-A71 vaccines have been developed in China (5,6). EV-A71 infections, along with other enterovirus serotypes belonging to the species Enterovirus A (EV-A) (7), are mainly associated with HFMD, which is characterized in children by a brief febrile illness and typical rash, with or without mouth ulcers (8). EV-A71 and coxsackievirus (CV) A16 were the most frequent serotypes involved in HFMD outbreaks throughout Asia during 1998–2010 (1,9). In the past 5 years, however, CV-A6 has emerged as a new important pathogen worldwide (10–19), and several studies have documented the more severe and extensive dermatologic presentations of CV-A6 HFMD (16,20–25). Surveillance of HFMD could lead to better detection of the upsurge of EV-A71 or another serotype associated with severe or distinct clinical features. In Western countries, surveillance of enterovirus infections is undertaken by virology laboratories and is thus restricted to enterovirus-infected persons admitted to hospitals (19,26). Children with HFMD or herpangina are usually evaluated and managed in ambulatory settings, and virologic investigations are rarely performed. Consequently, a clear gap exists in the knowledge of the epidemiology and clinical impact of HFMD and herpangina and of the enteroviruses involved in countries outside Asia.
We set up a local surveillance system run by pediatricians in ambulatory care settings that was effective in detecting HFMD outbreaks and the associated enterovirus serotypes (13). We have now extended this surveillance to cover the whole of France. The objectives of this study were to describe the epidemiology of enterovirus serotypes associated with HFMD and herpangina in France and to compare the clinical characteristics of HFMD and herpangina according to enterovirus serotypes.
Study Population and Design
The study was a 1-year prospective investigation of children with HFMD or herpangina who were seen by their pediatrician during April 2014–March 2015. The sentinel surveillance was performed by 47 pediatricians selected from among 118 volunteers by stratified sampling in different regions of France; 20 of the 22 French administrative regions were represented (Technical Appendix). Sentinel pediatricians were requested to collect throat or buccal swab specimens from children with clinically diagnosed HFMD/herpangina. HFMD was defined by the presence of >2 of the following signs: buccal or peribuccal ulcers; eruption on palms, soles, buttocks, knees, or elbows; or a generalized eruption. Herpangina was defined by the presence of oral ulcers predominating on the posterior part of the buccal cavity. A standardized case report form collected anonymized information on the patient’s demographics (e.g., birth date and sex); clinical signs at presentation, including fever, eruption type, and localization (e.g., palms, soles, buttocks, knees, elbows, lower limbs, upper limbs, generalized, or any other localization), buccal or peribuccal ulcers, gingivostomatitis, herpangina, and digestive/respiratory/ear, nose, and throat/neurologic signs; the onset date of the disease; and the date of sample collection. Environmental data (e.g., number of siblings, attendance at school or a daycare center, and ill contacts) were also recorded. Fever was defined as a rectal temperature >38°C. On the basis of the items checked, typical HFMD was defined as the presence of >2 of the following signs, as listed in the HFMD definition of the World Health Organization (8): oral ulcerations, eruption on palms, soles, buttocks, knees, or elbows. Clinical signs were considered to be atypical if eruption occurred at anatomic sites not listed in the World Health Organization HFMD definition or if it was generalized.
Written informed consent was obtained from the parents or guardians of all participants, none of whom received a stipend. The study was approved by the Ethics Review Committee of the University Hospital of Clermont-Ferrand, France (reference AU1098), and by the French National Agency for the Safety of Medicines and Health Products (reference 140021-B41).
Sample Collection
Throat or buccal specimens were collected with a flocked swab placed in a universal virus transport system (Copan Italia, s.p.a., Brescia, Italy). After the sampling, swabs were conserved at 2° –8°C and sent weekly to the National Reference Laboratory for Enterovirus and Parechovirus (Clermont-Ferrand, France) for enterovirus testing.
Diagnosis of Enterovirus Infection and Molecular Typing of Strains in Clinical Specimens
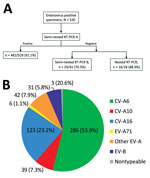
Viral RNA was extracted from 200 µL of the universal virus transport medium on the NucliSens easyMAG automated system (bioMérieux, Marcy l’Etoile, France) by the specific B protocol (elution volume 50 µL). Enterovirus diagnosis was performed by real-time reverse transcription PCR (RT-PCR) (Enterovirus R-gene, bioMérieux). Molecular typing (Figure 1, panel A) was first performed by a semi-nested RT-PCR with primers specifically developed for EV-A types (RT-PCR A) that targets the viral protein (VP) 3–VP1 coding region of the enterovirus genome. The first round of the RT-PCR EV-A assays were performed in a final volume of 25 µL with primers HEVAS1405 (5′-GGNTCNTTYATGGCNACNGGNAARATG-3′, location 1,405–1,531 relative to the genome of CV-A6 Gdula strain) and EVAR2C (5′-CGGTGYTTGCTCTTGAACTGCATG-3′, location 4,439–4,416) at a final concentration of 0.5 µmol/L, each by using the One-Step RT-PCR kit (QIAGEN, Courtaboeuf, France). The amplification program was as follows: 1 cycle of 30 min at 50°C; 1 cycle of 15 min at 95°C; 41 cycles of 30 s at 94°C, 50 s at 55°C, and 2 min at 72°C; and a final cycle of 10 min at 72°C. The second round RT-PCR A assays were performed in a final volume of 50 µL by using the Taq polymerase Kit (QIAGEN) and contained 5 µL of the first RT-PCR A amplicons and primers HEVAS1405 and HEVAR2429 (5′-GTNGGRTANCCRTCRTARAACC-3′, location 2,450–2,429) at a final concentration of 0.4 µmol/L. The reaction was run under the following conditions: 1 cycle of 2 min at 94°C; 39 cycles of 15 s at 94°C, 50 s at 54°C, and 50 s at 72°C; and a final cycle of 5 min at 72°C. If results were negative, a semi-nested RT-PCR was performed with primers specific for the species Enterovirus B (EV-B), HEVBS1695/EV2C (first round) and HEVS1695/HEVBR132 (second round) (27). The amplification programs were the same as those described except for the hybridization step, which was performed at 58°C. Alternatively, genotyping was attempted with nonspecies-specific primers to amplify the partial VP1 gene (28). Visible RT-PCR products after gel electrophoresis were purified and subjected to nucleotide sequencing with the same primers used for the amplification for the semi-nested RT-PCR A or, as previously described (27,28), by using the BigDye Terminator v1.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Ville-bon sur Yvette, France). The sequencing was performed on an ABI3500Dx genetic analyzer (Thermo Fisher Scientific). Virus identification was performed by BLAST analysis (http://www.ncbi.nlm.gov/BLAST) and confirmed by phylogenetic analysis (13). The results were prospectively sent to the attending pediatricians, and monthly feedback on the overall findings of the surveillance was provided.
Phylogenetic Analyses
To investigate the spatiotemporal relationships among virus variants, we compared the nucleotide VP1 sequences assigned to the enterovirus serotype CV-A6 with homologous sequences available in public databases. We discarded redundant sequences from the final alignment of 238 sequences of 369 nt (i.e., 159 sequences determined in this study and 79 publicly available sequences). The phylogenetic relationships were inferred using a Bayesian method implemented in the BEAST package version 1.8 (http://beast.bio.ed.ac.uk). The uncorrelated lognormal molecular clock was employed with a flexible Bayesian skyline plot coalescent prior and the general time reversible model of nucleotide substitution. The Markov chain Monte Carlo were run for 200 million generations. We calculated maximum clade credibility trees by using the Tree Annotator program version 1.5.4 in BEAST. Topological support was assessed by calculating the posterior probability (pp) density for each node. All sequences were deposited into the GenBank database (accession nos. LT595894–LT596052). To characterize the EV-A71 strains, we compared partial VP1 sequences with reference sequences for genogroups A–F and subgenogroups B0–B5 and C1–C5. Phylogenetic analysis was performed with the neighbor-joining method and the Tamura-Nei model of sequence evolution implemented in MEGA6 software (http://www.megasoftware.net).
Statistical Analyses
We performed statistical analyses with Stata 13 software (StataCorp LP, College Station, TX, USA). The tests were 2-sided, with a type I error set at α = 0.05. Patient characteristics were presented as mean (±SD) for continuous data (assumption of normality assessed by the Shapiro–Wilk test) and as the number of patients and associated percentages for categorical parameters. We classified patients according to statistical distribution and epidemiologic relevance into 4 age groups: 1) <1 year old, 2) >1 year old, 3) 2 to <3 years old, and 4) >3 years old. We compared the independent groups (i.e., age groups and CV-A6 infections [yes/no]) by χ2 or Fisher exact test for categorical variables and by analysis of variance (ANOVA) or Kruskall-Wallis test for quantitative parameters (assumption of homoscedasticity analyzed by Fisher-Snedecor test). When appropriate (ANOVA or Kruskall-Wallis; p<0.05), we performed post hoc tests (Tukey-Kramer after ANOVA and Dunn after Kruskall-Wallis test) for multiple comparisons, particularly for comparisons between classes of age. Multivariate analyses (logistic regression for dichotomous independent variable) were performed to take into account adjustment on covariates fixed according to univariate results and clinical relevance (i.e., age at enrolment and time between onset and consultation).
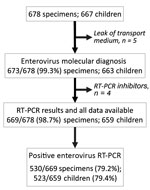
Of the 659 children enrolled in the study, 523 (79.3%) had an enterovirus infection. Ten patients experienced 2 episodes of HFMD/herpangina during the study period and had a specimen collected; 2 successive enterovirus infections associated with different serotypes occurred in 7 children at intervals of 3 weeks to 5.5 months. For the other 3 patients, only 1 episode was associated with an enterovirus infection.
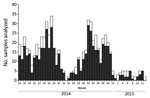
Overall, 669 specimens were analyzed, of which 530 (79.2%, 95% CI 75.9–82.2) tested positive for enterovirus (Figure 2). Mean patient enterovirus positivity rate for participating pediatricians was 75.1% (range 50.0%–88.9%). Enterovirus-associated HFMD/herpangina showed biannual peaks of activity in early summer (weeks 25–27) and in autumn (week 42) (Figure 3).
An enterovirus serotype was identified for 527/530 (99.4%) of proven infections. The most frequent EV-A serotype was CV-A6 (286/530, 53.9%) followed by CV-A16 (123/530, 23.2%), CV-A10 (39/530, 7.3%), CV-A4 (18, 3.3%), CV-A8 (16, 3.0%), CV-A2 (5, 0.9%), and CV-A5 (5, 0.9%); 1 infection was CV-A12 (Figure 1, panel B). Twelve EV-B serotypes were identified: echovirus 16 (E-16) and E-18 (5 each); E-11 and coxsackievirus B3 (CV-B3) (4 each); CV-B1, CV-B2, CV-B4, CV-A9, and E-6 (2 each); and E-3, E-5, and E-25 (1 each). CV-A6 was predominant during both epidemic waves. Six EV-A71 infections were detected, most associated with typical HFMD (5/6, 83.3%). One patient had generalized eruption. Fever was reported for only 2/6 patients. EV-A71 strains belonged to subgenotypes C2 (n = 5) and C4 (n = 1). The C4 strain was identified in a 3-year-old child from Guangzhou, China, who was on a visit to France (data not shown).
The mean age of enterovirus-infected children was 2.1 years (range 1 month–10.5 years). The highest rate of infections was observed in children 1–2 years of age. Fever was reported in (397/530, 74.9%) of enterovirus-infected children. Cutaneous eruption was observed in 456/530 (86%) children and affected, in decreasing order, the palms, soles, buttocks, and elbows. HFMD was the predominant clinical presentation (342/530, 64.5%). Herpangina was reported in 304/530 (57.4%) of cases and was associated with clinical signs of HFMD in 241/304 (79.2%). Lesions were also frequently observed on the limbs (188/530, 35.4%) and the face (perioral and earlobes, 161/530, 30.4%). Atypical HFMD was observed in 247/530 (46.6%) children (Table 1). The proportions of enterovirus-infected children were not significantly different between children presenting with typical HFMD (95/116, 81.9%) or atypical HFMD (247/281, 87.9%).
“Eczema coxsackium or herpeticum” was reported in 8 children, of whom 7 had a CV-A6 infection and 1 a CV-A10 infection. An eruption mimicking a Gianotti-Crosti syndrome caused by different enterovirus serotypes (CV-A6 [n = 3]; CV-A10 [n = 1]; or CV-A16 [n = 1]) was reported in 5 of these 8 children. Exposure to ill contacts was reported for 155/469 (33%) enterovirus-infected children with available environmental data, a proportion significantly higher than that among non–enterovirus-infected children (25/127, 19.7%; p = 0.004).
Patients with CV-A6 HFMD/herpangina were significantly younger than patients with other EV-A serotypes (p<0.001) (Table 1). The clinical features of CV-A6-associated HFMD were also significantly different. Atypical HFMD was more frequently reported in CV-A6-infected children (181/286, 63.3%) than in children infected with other enterovirus serotypes: CV-A16 (42/123, 34.1%), CV-A10 (14/39, 35.8%), CV-A4 and E-16 (2 cases each), and CV-A12, E-6, E-11, E18, or CV-B4 (1 case each). Typical HFMD or herpangina alone were significantly more frequent in children infected by the other EV-A serotypes (p<0.001) (Table 1). The most frequent serotypes associated with typical HFMD were CV-A16 (54/95, 56.8%) and CV-A6 (32/95, 33.7%), followed by EV-A71 (n = 4), CV-A8 and CV-A10 (n = 2 each), and E-5 (n = 1). The most frequent serotypes associated with herpangina alone were CV-A6 (14/63, 22.2%), CV-A4 (11/63, 17.4%), CV-A8 (10/63, 15.9%), and CV-A10 (8/63, 12.7%). The differences between CV-A6 infections and non–CV-A6 infections remained statistically significant by multivariate analyses.
Atypical HFMD was less frequent in patients <1 year of age than in children 1–2 and >3 years of age. Herpangina alone was more frequent in children <1 year of age than in older children (Table 2).
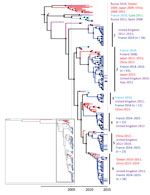
The CV-A6 strains sampled in France in 2014 and 2015 were grouped in 6 co-circulating lineages supported by high posterior probability values (Figure 4). The nucleotide identities within lineages ranged from 95.3% to 98.9% (98.3%–99.5% amino acid identities). Between lineages, the nucleotide identities ranged from 91.9% to 96.3% (97.5%–98.4% amino acid identities), the highest divergence being observed between lineages 1 and 2 and the lowest between 5 and 6. In lineage 1, the virus strains collected in France were temporally distantly related to viruses collected 1–3 years earlier in the United Kingdom. In the other 5 lineages, the CV-A6 virus strains sampled in France displayed close temporal relationships to viruses recovered in other countries in Europe since 2010. In the lineages 2 and 6, close genetic and temporal relationships were also estimated between virus strains recovered in France and countries in Asia. The 2014 CV-A6 viruses in lineage 3 were genetically related to those recovered in 2010 in France.
This prospective ambulatory clinic–based surveillance of HFMD/herpangina in children revealed the global effect of these diseases in France. Data were collected from a standardized report of clinical signs, which provided a comprehensive description of the clinical characteristics of these syndromes associated with different enterovirus serotypes. During April 2014–March 2015, CV-A6 infections were associated with HFMD in 74.4% cases and herpangina in 57.7% of cases. These proportions were inverted during the 2010 HFMD outbreak in central France: 50% for HFMD cases and 70% for herpangina (13). In addition, the dermatologic presentation of CV-A6 HFMD cases was more frequently unusual (63.3%), with eruptions extending beyond the typical sites of HFMD (i.e., soles, palms, buttocks, and knees or elbows) than in HFMD cases caused by other enterovirus serotypes. Our findings suggest that the clinical presentation of CV-A6 infections in France shifted to atypical HFMD during 2010–2014, as observed in China during 2008–2013 (18). In our study, the co-circulation of 6 virus lineages in 2014 is consistent with the hypothesis of multiple introductions of genetically distinct CV-A6 strains. In addition, a genetic analysis of complete CV-A6 genomes showed that strains collected during the 2012–2013 outbreak in Shanghai, China, were recombinant compared with strains collected before 2009 and were more frequently associated with generalized rash (29).
Comparative analyses of whole virus genomes should be expanded on large sequence data derived from prospective epidemiologic studies to investigate whether the changes in the clinical features of CV-A6 infections reported here are determined by viral factors. The relationship between CV-A6 and atypical HFMD has been reported in earlier studies that described frequent unusual morphology or extent of cutaneous findings (15,20–25,30), such as “eczema coxsackium,” Gianotti-Crosti-like eruption, and purpuric eruption (21). However, these studies were either retrospective, focused on severe or atypical HFMD, or performed in dermatology pediatric centers, which might have biased the clinical spectrum of CV-A6 HFMD cases toward more severe or atypical presentations. Although we cannot exclude the possibility that pediatricians were more prone to include children with unusual presentations of HFMD, our study confirms that CV-A6 is more frequently associated with atypical HFMD even in an ambulatory setting.
Of note, atypical HFMD was reported in 66/244 (27%) of non–CV-A6-associated HFMD cases. This result might be attributable to the definition of atypical HFMD we used, which was, in contrast to that of typical HFMD (8), the involvement of a nontypical anatomic site for HFMD. The lack of a consensus definition of atypical HFMD and the fact that the collected clinical data vary between studies hamper rigorous comparisons between them. Further investigations based on prospective ambulatory clinic–based surveillance of HFMD are needed to determine whether our observation is attributable to a specific increase in the circulation of CV-A6 or to a global change in the transmission of enterovirus strains. The documentation of unusual presentations of HFMD by enterovirus genotyping is useful for detecting the emergence of a new serotype with distinct clinical features.
The clinical courses of typical and atypical HFMD seemed similar in our cohort because no complications were reported. The extensive or unusual nature of the cutaneous manifestations of CV-A6 HFMD does not seem to increase the risk for severe systemic illness (21). Although to a lesser extent than with EV-A71, severe CV-A6 HFMD cases, defined by the presence of neurologic signs (e.g., meningitis, encephalitis, acute flaccid paralysis, and seizures) or cardiopulmonary signs, have been reported, with a frequency ranging from 3.6 % to 18.2% during the recent CV-A6 outbreaks in China (18,31–34). Meningitis rather than encephalitis was more frequently associated with severe CV-A6 HFMD (32,33). However, clinicians should be aware of the potential neurotropism of all EVs. As exemplified by EV-A71 outbreaks (35) and more recently EV-D68 outbreaks (2), the high rate of circulation of CV-A6, either symptomatic or asymptomatic, can lead to the more frequent observation of this serotype in association with neurologic signs.
As is the case with many common and self-limited illnesses, the children in our study might not have attended all medical consultations, thus rendering the surveillance incomplete. The comprehensive recruitment of all children with HFMD/herpangina is time-consuming and not feasible in the routine practice of ambulatory pediatrics, which might have resulted in a lower inclusion rate. We do not have recorded long-term follow-up or the specific CV-A6–associated clinical entities described by Mathes et al. (13), and we were not able to assess the occurrence of onychomadesis or desquamation of the extremities, which have been frequently associated with CV-A6 outbreaks (9–11,15,20,32).
This study contributes to a more comprehensive view of the epidemiology of HFMD/herpangina in France and the clinical spectrum of HFMD/herpangina associated with enterovirus, in particular with CV-A6. The often unusual presentation of HFMD can be challenging for clinicians, and this study might therefore help improve the differential diagnosis of HFMD by primary care physicians and the detection of future HFMD outbreaks. Syndromic surveillance of HFMD/herpangina by pediatricians in ambulatory setting with prospective and standardized collection of clinical data in combination with enterovirus testing and genotyping are useful for monitoring the epidemiology of enterovirus infections, for the timely detection of peaks of highest activity, and for determining the enterovirus serotypes involved, leading to better detection of outbreaks associated with EV-A71 or any other serotype associated with severe or distinct clinical features.
Dr. Mirand is a clinical researcher in virology at the University Hospital in Clermont-Ferrand and the National Reference Laboratory of Enteroviruses and Parechoviruses. Her research interest is the epidemiology of enterovirus infections, with a focus on enteroviruses A associated with hand, foot and mouth disease.
Acknowledgments
This study was made possible by the close involvement of all ambulatory pediatricians who took part in the sentinel surveillance system on behalf of the Association Française de Pédiatrie Ambulatoire (http://www.afpa.org): Anne-Sophie Belloin-Perraud, Pierre Blanc, Patrice Bouissou, Marie-Annick Burgess, Marie-Edith Burthey, Michelle Carsenti, Marie-Christine Chauvel, Caroline Colombet, François Corrard, Liliane Cret, Veronique Dagrenat-Taleb, Simon Dib, Philippe Franchineau, Cécile Guiheneuf, Jean-Louis Guillon, Sylvie Hubinois, Pierre Jarry, Flaviane Kampf-Maupu, Fabienne Kochert, Caroline Laffort, Anne-Louise Lambert, Francine Lecailler, Hélène Le Scornet, Murielle Louvel, Nicolas Mathieu, Jean-François Maurer, Bruno Mazauric, Agnès Mercier, Frédéric Mestre, Solange Moore-Wipf, Sylvie Pauliat-Desbordes, Florence Pichot-Jallas, Saholy Razafinarivo Schoreisz, Céline Rollet-Lautraite, Sylvie Sandid, Isabelle Sartelet, Simone Saumureau, Philippe Simon, Abderrahim Terrache, Georges Thiebault, Jean-Pierre Tiberghien, Eric Van Melkebeke, Sandrine Vernoux, Martine Wagner-Vaucard, and Andreas Werner. These pediatricians were involved in the recruitment of patients and did not receive any compensation. We are also grateful to the children and families who made this study possible. We also thank the virologists Jean-Michel Mansuy and Catherine Mengelle for allowing us to include sequences of CV-A6 strains from hospitalized patients at the University Hospital of Toulouse, France. We are grateful to Nathalie Rodde and Emilie Leroy for excellent technical assistance in enterovirus testing and genotyping. We thank Jeffrey Watts for revision of the English manuscript.
The Centre National de Référence des Enterovirus-Parechovirus is supported by an annual grant from the French national public health network (Santé Publique France).
References
- Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–90.DOIPubMedGoogle Scholar
- Messacar K, Schreiner TL, Maloney JA, Wallace A, Ludke J, Oberste MS, A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet. 2015;385:1662–71.DOIPubMedGoogle Scholar
- World Health Organization. Enterovirus surveillance guidelines: guidelines for enterovirus surveillance in support of the Polio Eradication Initiative. Geneva: The Organization; 2015 [cited 2015 Apr 2]. http://www.euro.who.int/en/publications/abstracts/enterovirus-surveillance-guidelines.-guidelines-for-enterovirus-surveillance-in-support-of-the-polio-eradication-initiative
- Shaw J, Welch TR, Milstone AM. The role of syndromic surveillance in directing the public health response to the enterovirus D68 epidemic. JAMA Pediatr. 2014;168:981–2.DOIPubMedGoogle Scholar
- Zhu F, Xu W, Xia J, Liang Z, Liu Y, Zhang X, Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. N Engl J Med. 2014;370:818–28.DOIPubMedGoogle Scholar
- Mao QY, Wang Y, Bian L, Xu M, Liang Z. EV71 vaccine, a new tool to control outbreaks of hand, foot and mouth disease (HFMD). Expert Rev Vaccines. 2016;15:599–606.DOIPubMedGoogle Scholar
- Adams MJ, Lefkowitz EJ, King AMQ, Bamford DH, Breitbart M, Davison AJ, Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2015). Arch Virol. 2015;160:1837–50.DOIPubMedGoogle Scholar
- World Health Organization. A guide to clinical management and public health response for hand, foot and mouth disease. Geneva: The Organization; 2011 [cited 2015 Apr 2]. http://www.wpro.who.int/emerging_diseases/documents/HFMDGuidance/en
- Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis. 2014;14:308–18.DOIPubMedGoogle Scholar
- Österback R, Vuorinen T, Linna M, Susi P, Hyypiä T, Waris M. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis. 2009;15:1485–8.DOIPubMedGoogle Scholar
- Wu Y, Yeo A, Phoon MC, Tan EL, Poh CL, Quak SH, The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis. 2010;14:e1076–81.DOIPubMedGoogle Scholar
- Wei SH, Huang YP, Liu MC, Tsou TP, Lin HC, Lin TL, An outbreak of coxsackievirus A6 hand, foot, and mouth disease associated with onychomadesis in Taiwan, 2010. BMC Infect Dis. 2011;11:346.DOIPubMedGoogle Scholar
- Mirand A, Henquell C, Archimbaud C, Ughetto S, Antona D, Bailly JL, Outbreak of hand, foot and mouth disease/herpangina associated with coxsackievirus A6 and A10 infections in 2010, France: a large citywide, prospective observational study. Clin Microbiol Infect. 2012;18:E110–8.DOIPubMedGoogle Scholar
- Cabrerizo M, Tarragó D, Muñoz-Almagro C, Del Amo E, Domínguez-Gil M, Eiros JM, Molecular epidemiology of enterovirus 71, coxsackievirus A16 and A6 associated with hand, foot and mouth disease in Spain. Clin Microbiol Infect. 2014;20:O150–6.DOIPubMedGoogle Scholar
- Fujimoto T, Iizuka S, Enomoto M, Abe K, Yamashita K, Hanaoka N, Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis. 2012;18:337–9.DOIPubMedGoogle Scholar
- Sinclair C, Gaunt E, Simmonds P, Broomfield D, Nwafor N, Wellington L, Atypical hand, foot, and mouth disease associated with coxsackievirus A6 infection, Edinburgh, United Kingdom, January to February 2014. Euro Surveill. 2014;19:20745.DOIPubMedGoogle Scholar
- Hayman R, Shepherd M, Tarring C, Best E. Outbreak of variant hand-foot-and-mouth disease caused by coxsackievirus A6 in Auckland, New Zealand. J Paediatr Child Health. 2014;50:751–5.DOIPubMedGoogle Scholar
- Zeng H, Lu J, Zheng H, Yi L, Guo X, Liu L, The epidemiological study of coxsackievirus A6 revealing hand, foot and mouth disease epidemic patterns in Guangdong, China. Sci Rep. 2015;5:10550.DOIPubMedGoogle Scholar
- Abedi GR, Watson JT, Pham H, Nix WA, Oberste MS, Gerber SI. Enterovirus and human parechovirus surveillance—United States 2009–2013. MMWR Morb Mortal Wkly Rep. 2015;64:940–3.DOIPubMedGoogle Scholar
- Flett K, Youngster I, Huang J, McAdam A, Sandora TJ, Rennick M, Hand, foot, and mouth disease caused by coxsackievirus a6. Emerg Infect Dis. 2012;18:1702–4.DOIPubMedGoogle Scholar
- Mathes EF, Oza V, Frieden IJ, Cordoro KM, Yagi S, Howard R, “Eczema coxsackium” and unusual cutaneous findings in an enterovirus outbreak. Pediatrics. 2013;132:e149–57.DOIPubMedGoogle Scholar
- Huang WC, Huang LM, Lu CY, Cheng AL, Chang LY. Atypical hand-foot-mouth disease in children: a hospital-based prospective cohort study. Virol J. 2013;10:209.DOIPubMedGoogle Scholar
- Chung WH, Shih SR, Chang CF, Lin TY, Huang YC, Chang SC, Clinicopathologic analysis of coxsackievirus a6 new variant induced widespread mucocutaneous bullous reactions mimicking severe cutaneous adverse reactions. J Infect Dis. 2013;208:1968–78.DOIPubMedGoogle Scholar
- Feder HM Jr, Bennett N, Modlin JF. Atypical hand, foot, and mouth disease: a vesiculobullous eruption caused by Coxsackie virus A6. Lancet Infect Dis. 2014;14:83–6.DOIPubMedGoogle Scholar
- Hubiche T, Schuffenecker I, Boralevi F, Léauté-Labrèze C, Bornebusch L, Chiaverini C, ; Clinical Research Group of the French Society of Pediatric Dermatology Groupe de Recherche Clinique de la Société Française de Dermatologie Pédiatrique. Dermatological spectrum of hand, foot and mouth disease from classical to generalized exanthema. Pediatr Infect Dis J. 2014;33:e92–8.DOIPubMedGoogle Scholar
- Antona D, Lévêque N, Chomel JJ, Dubrou S, Lévy-Bruhl D, Lina B. Surveillance of enteroviruses in France, 2000-2004. Eur J Clin Microbiol Infect Dis. 2007;26:403–12.DOIPubMedGoogle Scholar
- Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–704.DOIPubMedGoogle Scholar
- Mirand A, Henquell C, Archimbaud C, Chambon M, Charbonne F, Peigue-Lafeuille H, Prospective identification of enteroviruses involved in meningitis in 2006 through direct genotyping in cerebrospinal fluid. J Clin Microbiol. 2008;46:87–96.DOIPubMedGoogle Scholar
- Feng X, Guan W, Guo Y, Yu H, Zhang X, Cheng R, A novel recombinant lineage’s contribution to the outbreak of coxsackievirus A6-associated hand, foot and mouth disease in Shanghai, China, 2012-2013. Sci Rep. 2015;5:11700.DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention (CDC). Notes from the field: severe hand, foot, and mouth disease associated with coxsackievirus A6 - Alabama, Connecticut, California, and Nevada, November 2011-February 2012. MMWR Morb Mortal Wkly Rep. 2012;61:213–4.PubMedGoogle Scholar
- Li JL, Yuan J, Yang F, Wu ZQ, Hu YF, Xue Y, Epidemic characteristics of hand, foot, and mouth disease in southern China, 2013: coxsackievirus A6 has emerged as the predominant causative agent. J Infect. 2014;69:299–303.DOIPubMedGoogle Scholar
- Hongyan G, Chengjie M, Qiaozhi Y, Wenhao H, Juan L, Lin P, Hand, foot and mouth disease caused by coxsackievirus A6, Beijing, 2013. Pediatr Infect Dis J. 2014;33:1302–3.DOIPubMedGoogle Scholar
- Yang F, Yuan J, Wang X, Li J, Du J, Su H, Severe hand, foot, and mouth disease and coxsackievirus A6-Shenzhen, China. Clin Infect Dis. 2014;59:1504–5.DOIPubMedGoogle Scholar
- Lu QB, Zhang XA, Wo Y, Xu HM, Li XJ, Wang XJ, Circulation of Coxsackievirus A10 and A6 in hand-foot-mouth disease in China, 2009-2011. PLoS One. 2012;7:e52073.DOIPubMedGoogle Scholar
- Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–105.DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 22, Number 11—November 2016
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Audrey Mirand, CHU Clermont-Ferrand, Laboratoire de Virologie, CNR des Entérovirus et Parechovirus Laboratoire Associé, 63003 Clermont-Ferrand CEDEX, France
Top