Volume 22, Number 6—June 2016
Dispatch
Whole-Genome Analysis of Cryptococcus gattii, Southeastern United States
Figure 2
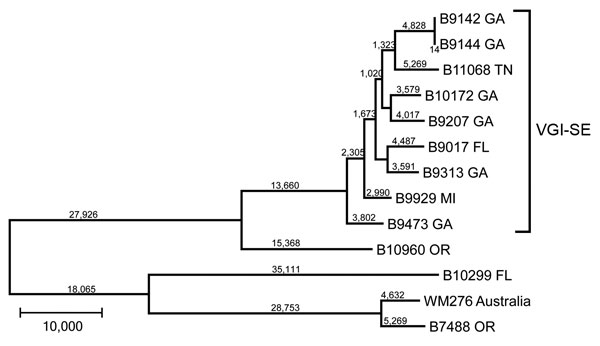
Figure 2. Maximum-parsimony tree of whole-genome sequence data of isolates of Cryptococcus gattii from the southeastern United States. All bootstrap values were 100%. Numbers on branches are SNPs. Nearest neighbor isolates were included for comparison, and an environmental VGI isolate from Australia was used as an outgroup. SNP, single-nucleotide polymorphism; VGI-SE, VGI southeastern clade. Scale bar indicates 10,000 SNPs.
Page created: May 16, 2016
Page updated: May 16, 2016
Page reviewed: May 16, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.