Volume 23, Number 10—October 2017
Dispatch
Berlin Squirrelpox Virus, a New Poxvirus in Red Squirrels, Berlin, Germany
Abstract
Near Berlin, Germany, several juvenile red squirrels (Sciurus vulgaris) were found with moist, crusty skin lesions. Histology, electron microscopy, and cell culture isolation revealed an orthopoxvirus-like infection. Subsequent PCR and genome analysis identified a new poxvirus (Berlin squirrelpox virus) that could not be assigned to any known poxvirus genera.
The Eurasian red squirrel (Sciurus vulgaris) is the only species of tree squirrels endemic throughout most of Europe. Although they are usually abundant, red squirrels are endangered or extinct in some regions in Great Britain and Ireland that are co-inhabited by invasive eastern gray squirrels (Sciurus carolinensis), which were introduced from North America in the late 19th century. One major threat is the transmission of squirrelpox virus (SQPV) from the gray squirrel reservoir host to red squirrels, which succumb to lethal infections (1). SQPV had been assigned to the parapoxviruses due to morphological similarities (2), but the latest viral genome data placed it in a separate clade within the poxvirus family (3). Recently, different poxviruses have been associated with similar lesions in American red squirrels (Tamiasciurus hudsonicus) from Canada (4), but except for a single case report from Spain (5), no poxvirus infections in squirrels have been reported in continental Europe.
In 2015 and 2016, at least 10 abandoned weak juvenile red squirrels were submitted to a sanctuary near Berlin, Germany. The animals had exudative and erosive-to-ulcerative dermatitis with serocellular crusts at auricles, noses, digits, tails, and genital/perianal regions. Skin specimens from affected animals were investigated by electron microscopy (EM) and PCR. Three animals that died under care were submitted for necropsy. We obtained samples of all organs for histological and PCR examination. We used 1 sample of a skin lesion for virus propagation in cell culture.
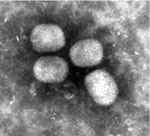
EM-negative staining of skin lesions from all animals led to the discovery of brick-shaped poxvirus particles with irregular threadlike surface fibers and an average size of 294 nm × 221 nm (Figure 1). Pathological findings of corresponding skin lesions were consistent with poxvirus infection (ballooning degeneration of epidermal keratinocytes, numerous intracytoplasmic inclusion bodies, epidermal ulceration with suppurative inflammation, and secondary bacterial infection). All inner organs had either no pathological changes or lesions unrelated to poxvirus infection.
To confirm the morphologic diagnosis, we extracted DNA from skin lesions and performed various PCRs. An orthopoxvirus (OPV)–specific PCR showed negative results (6); a parapoxvirus (PPV)–specific PCR (6), a leporipoxvirus-specific PCR (A. Nitsche and L. Schrick, unpub. data), and a poxvirus-screening PCR (7) were positive for some samples. Obtained sequence fragments indicated poxviral relatedness but did not allow for the assignment to a poxvirus genus. Thus, we performed massively parallel sequencing. We directly subjected DNA extracted from a skin lesion on the foot of a dead animal to Nextera XT Library preparation and sequenced it on an Illumina HiSeq 1500 instrument (Illumina, San Diego, CA, USA), yielding 7,242,301 paired-end reads (150 + 150 bases, rapid run mode). Mapping (8) the obtained reads to all poxvirus reference sequences available in GenBank in high-sensitivity mode provided no notable results, which pointed to a virus with a highly deviant genome. Therefore, we separated poxviral reads from background data using RAMBO-K version 1.2 (9) and assembled the resulting 1,520,811 reads (10), yielding 1 single contig of 142,974 bp with ≈460-fold coverage after manual iterative mapping and scaffolding. We confirmed the genomic sequence by resequencing (Illumina MiSeq) of a Vero E6 cell-culture isolate obtained from a different skin specimen of the same animal. We named the new virus Berlin SQPV (BerSQPV), and uploaded the combined sequence information to GenBank (accession no. MF503315). Direct sequencing of DNA from skin samples of three other animals from the same origin yielded sequences with >99.9% identity to BerSQPV.
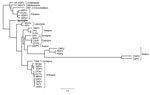
We compared characteristics of BerSQPV to related viruses and found that the EM structure shows features typical for OPV but the genome size of ≈143 kb is more consistent with PPV or SQPV from the United Kingdom (11) than with the large genome of OPV, whereas the guanine-cytosine (GC) content of 38.5% is more consistent with OPV and leporipoxvirus than with PPV and SQPV from the United Kingdom. Therefore, we explored the genomic relationship of BerSQPV to other chordopoxviruses. Pairwise alignments of each of the chordopoxvirus genomes available in GenBank with the BerSQPV genome resulted in a pairwise identity of at most 47% to tanapox virus isolate TPV-Kenya (accession no. EF420156.1). The retrieved phylogenetic tree (Figure 2) demonstrates that BerSQPV cannot be assigned to any of the known poxvirus genera; moreover, it does not cluster with the only other squirrel poxvirus with a published genome sequence (11). Further phylogenetic analyses based on conserved single genes frequently used for poxvirus tree calculations (A3L, F10L+F12L, F13L, E13L, E9L [VACV Copenhagen nomenclature]) showed similar results (A56R was not used for tree calculations because this open reading frame is too divergent among the Chordopoxvirinae), with BerSQPV forming a unique branch (data not shown). In addition, any partial sequences of SQPV available in GenBank were aligned to BerSQPV, showing a maximal sequence identity of 64.3% to gene E9L (GenBank accession no. AY340976.1), further emphasizing the uniqueness of this newly identified virus.
We designed a BerSQPV-specific quantitative PCR based on the genome sequence as a tool for future investigations (primer BerSQPV_F: ggAAgTTTTCCCATACCAACTgA, primer BerSQPV_R: ATCTCAAACCgCAgACggTA, probe BerSQPV_TM: FAM-ACTgTTATTCTTAgCgTAATT). Sensitivity was <10 genome equivalents per reaction amplifying plasmid dilution rows. We first validated the specificity in silico during the design process, revealing the highest identity of 88% to cowpox virus Kostroma (GenBank accession no. KY369926.1), with mismatches in crucial positions in the primer and probe binding sites. Squirrel poxvirus strain Red squirrel UK (GenBank accession no. NC_022563.1) showed only 84% identity, with additional mismatches in amplification-relevant positions. Practical PCR testing using DNA from cowpox, monkeypox, ectromelia, parapox-ORF, myxoma, avipox, and molluscipox viruses showed no cross-reactivity.
The new specific quantitative PCR was subsequently applied to DNA from skin lesions archived from 1 squirrel found dead in 2014 in the Berlin area, 2 live squirrels from 2015, and 5 live squirrels from 2016, as well as various organs from 3 affected squirrels necropsied in 2015 (Table). Organ tissues yielded high BerSQPV DNA loads in the affected skin but low viral DNA loads for inner organs, findings in concordance with pathological findings, indicating the detection of viral DNA in the blood homogenously distributed throughout the organs with specific tropism for the skin. Low virus loads in inner organs are usually observed in poxvirus infections that do not generalize. PCR results indicate that this virus has been circulating in the Berlin area over the past 10 years.
We describe a new poxvirus, BerSQPV, isolated from red squirrels in Berlin, Germany, that causes pathological changes consistent with other epidermal poxvirus infections. Genome analysis revealed a unique sequence within the poxvirus family, as BerSQPV is not clustering to other poxvirus genera, including UK SQPV from red squirrels from Great Britain. In contrast to UK SQPV, which resembles PPV ultrastructurally (2), the ultrastructure of BerSQPV is comparable to that of OPV. Two other poxviruses from tree squirrels with ultrastructural appearance similar to BerSQPV have been reported: a Eurasian red squirrel from Spain with epidermal poxvirus lesions (5) and American red squirrels from Canada (15). Although no sequence information is available for the SQPV from Spain, the partial sequence analysis of SQPV from Canada showed the virus to also be distinct from all known mammalian poxviruses but most closely related to PPV, followed by UK SQPV (15).
BerSQPV is suspected to have been circulating for several years among Eurasian red squirrels in the greater Berlin area. Although diseased animals in care were handled in close contact, caretakers have remained asymptomatic, suggesting a negligible risk for human infection. Further detailed characterization of the isolated virus is ongoing.
Dr. Wibbelt is a senior veterinary pathologist at the Leibniz Institute for Zoo and Wildlife Research, Berlin, Germany. Her research interests include infectious diseases in wildlife animals, particularly small mammals. Mr. Tausch is a PhD student in bioinformatics at ZBS 1, Robert Koch Institute. His research interests include (meta)genomics with a focus on viruses, combining both application and development of algorithms.
Acknowledgment
The authors are grateful to Tanya Lenn, who encountered squirrels with suspicious lesions for many years, for providing samples. Special thanks are given to Dagmar Viertel, Zoltan Mezoe, Doris Krumnow, and Anja Ostrowski and the technicians of the German Consultant Lab for Poxviruses (RKI) for their excellent technical assistance, as well as to Ursula Erikli for copyediting.
References
- Sainsbury AW, Deaville R, Lawson B, Cooley WA, Farelly SS, Stack MJ, et al. Poxviral disease in red squirrels Sciurus vulgaris in the UK: spatial and temporal trends of an emerging threat. EcoHealth. 2008;5:305–16. DOIPubMedGoogle Scholar
- Scott AC, Keymer IF, Labram J. Parapoxvirus infection of the red squirrel (Sciurus vulgaris). Vet Rec. 1981;109:202. DOIPubMedGoogle Scholar
- McInnes CJ, Wood AR, Thomas K, Sainsbury AW, Gurnell J, Dein FJ, et al. Genomic characterization of a novel poxvirus contributing to the decline of the red squirrel (Sciurus vulgaris) in the UK. J Gen Virol. 2006;87:2115–25. DOIPubMedGoogle Scholar
- Himsworth CG, Musil KM, Bryan L, Hill JE. Poxvirus infection in an American red squirrel (Tamiasciurus hudsonicus) from northwestern Canada. J Wildl Dis. 2009;45:1143–9. DOIPubMedGoogle Scholar
- Obon E, Juan-Sallés C, McInnes CJ, Everest DJ. Poxvirus identified in a red squirrel (Sciurus vulgaris) from Spain. Vet Rec. 2011;168:86. DOIPubMedGoogle Scholar
- Kurth A, Nitsche A. Detection of human-pathogenic poxviruses. Methods Mol Biol. 2010;665:257–78. DOIPubMedGoogle Scholar
- Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–76. DOIPubMedGoogle Scholar
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. DOIPubMedGoogle Scholar
- Tausch SH, Renard BY, Nitsche A, Dabrowski PW. RAMBO-K: rapid and sensitive removal of background sequences from next generation sequencing data. PLoS One. 2015;10:e0137896. DOIPubMedGoogle Scholar
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. DOIPubMedGoogle Scholar
- Darby AC, McInnes CJ, Kjær KH, Wood AR, Hughes M, Martensen PM, et al. Novel host-related virulence factors are encoded by squirrelpox virus, the main causative agent of epidemic disease in red squirrels in the UK. PLoS One. 2014;9:e96439. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Himsworth CG, McInnes CJ, Coulter L, Everest DJ, Hill JE. Characterization of a novel poxvirus in a North American red squirrel (Tamiasciurus hudsonicus). J Wildl Dis. 2013;49:173–9. DOIPubMedGoogle Scholar
Figures
Table
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 23, Number 10—October 2017
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Andreas Nitsche, Centre for Biological Threats and Special Pathogens 1—Highly Pathogenic Viruses, Robert Koch Institute, Seestr 10, 13353 Berlin, Germany
Top