Volume 23, Number 7—July 2017
Dispatch
Locally Acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013
Figure
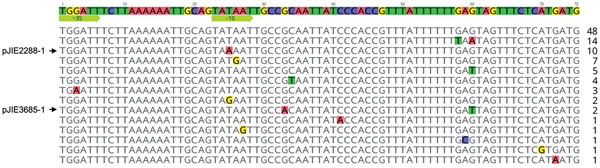
Figure. Differences in promoter and ribosome binding site regions of mcr-1 in plasmids from Escherichia coli in Australia (indicated by arrows) and in other sequences available from GenBank. The sequences end with the ATG start codon of mcr-1 and a second ATG codon that follows it. The −35 and −10 regions of the proposed promoter (11) are indicated by arrows. The numbers to the right indicate how many times each variant has been seen among available sequences.
References
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. DOIPubMedGoogle Scholar
- Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 2016;60:6973–6. DOIPubMedGoogle Scholar
- Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71:2066–70. DOIPubMedGoogle Scholar
- Valenzuela JK, Thomas L, Partridge SR, van der Reijden T, Dijkshoorn L, Iredell J. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J Clin Microbiol. 2007;45:453–60. DOIPubMedGoogle Scholar
- Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16:288–9. DOIPubMedGoogle Scholar
- Olaitan AO, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Khounsy S, et al. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J Antimicrob Chemother. 2015;70:3402–4.PubMedGoogle Scholar
- Zhang XF, Doi Y, Huang X, Li HY, Zhong LL, Zeng KJ, et al. Possible transmission of mcr-1–harboring Escherichia coli between companion animals and human. Emerg Infect Dis. 2016;22:1679–81. DOIPubMedGoogle Scholar
- Brouwer MS, Tagg KA, Mevius DJ, Iredell JR, Bossers A, Smith HE, et al. IncI shufflons: Assembly issues in the next-generation sequencing era. Plasmid. 2015;80:111–7. DOIPubMedGoogle Scholar
- Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis. 2016;16:284–5. DOIPubMedGoogle Scholar
- Yang YQ, Li YX, Song T, Yang YX, Jiang W, Zhang AY, et al. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother. 2017;61:e01201–61. DOIPubMedGoogle Scholar
- Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of MCR-1–producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 2016;60:4394–7. DOIPubMedGoogle Scholar
- Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. DOIPubMedGoogle Scholar
- Nguyen NT, Nguyen HM, Nguyen CV, Nguyen TV, Nguyen MT, Thai HQ, et al. Use of colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and its association with resistance in commensal Escherichia coli bacteria. Appl Environ Microbiol. 2016;82:3727–35. DOIPubMedGoogle Scholar
- Australian Commission on Safety and Quality in Health Care. AURA 2016: first Australian report on antimicrobial use and resistance in human health. Sydney: the Commission; 2016.
Page created: June 19, 2017
Page updated: June 19, 2017
Page reviewed: June 19, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.