Volume 24, Number 1—January 2018
Research Letter
Yellow Fever Virus DNA in Urine and Semen of Convalescent Patient, Brazil
Figure
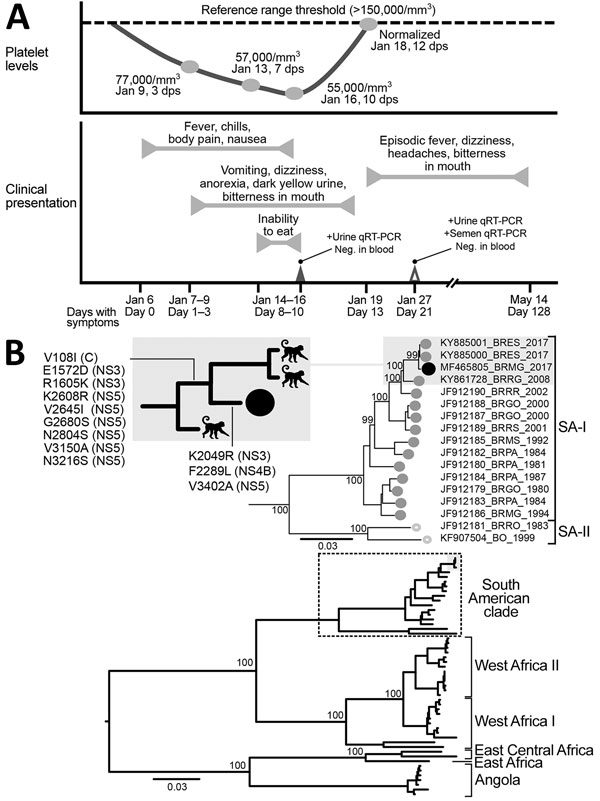
Figure. Clinical progression and detection of YFV RNA in urine and semen of convalescent patient, Brazil. A) Platelet levels, clinical parameters and symptoms, and test results over a 128-day period after initial symptoms were observed. B) Maximum-likelihood tree (midpoint-rooted) inferred by using complete genomes of YFV to distinguish major virus genotypes; dashed box indicates South American clade strains, enlarged at top. Numbers near nodes indicate percent bootstrap values after 10,000 replicates for major branches. Black circles indicate virus isolated in this study. Shaded boxes indicate monkey-derived virus sister taxa sampled during the same outbreak; inset at top left shows most parsimonious reconstructions of synapomorphic changes detected NS3, NS4B, and NS5 genes. GenBank accession number, geographic location code, and year of isolation are shown for virus isolates. Scale bars indicate nucleotide substitutions per site. C, capsid; dps, days postsymptom onset; Neg., negative; NS, nonstructural; qRT-PCR, quantitative reverse transcription PCR; SA, South America clade; YFV, yellow fever virus; +, positive.
1These authors contributed equally to this article.
2Current affiliation: Pirbright Institute, Pirbright, UK.