Volume 24, Number 12—December 2018
Research
Emerging Multidrug-Resistant Hybrid Pathotype Shiga Toxin–Producing Escherichia coli O80 and Related Strains of Clonal Complex 165, Europe
Figure 2
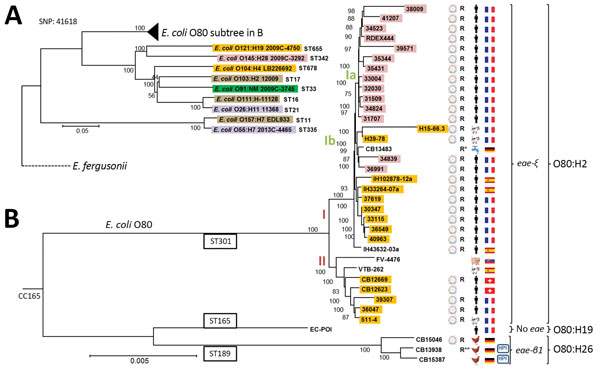
Figure 2. Phylogeny of 36 Escherichia coli serogroup O80 strains isolated from various sources and countries in Europe during 1998–2016 and their relationship to major enterohemorrhagic E. coli lineages. General phylogenic tree rooted on E. fergusonii (GenBank accession no. NC_011740), showing (A) the position of O80 strains among major enterohemorrhagic E. coli lineages (O157:H7 EDL933 NC_002655.2, O26:H11 11368 NC_013361.1, O111:H- 11128 NC_013364.1, O103:H2 12009 NC_013353.1, O55:H7 2013C-4465 CP015241, O91:NM 2009C-3745 JHGW00000000, O104:H4 LB226692 EO104H4LB.1, O145:H28 2009C-3292 JHHD00000000, and O121:H19 2009C-4750 JHGL00000000) and (B) a focused view of clonal complex 165, including O80 strains. The highlighted strains carry the Shiga toxin genes; the subtype of Stx is indicated by a color code as follows: purple, stx1a; yellow, stx2a; pink, stx2dactivable; brown, stx1a2a; dark green, stx1a2b. The presence of the pS88 like–plasmid is represented by using a plasmid scheme next to the strain number. R to the right of the plasmid indicates that the strain possesses >2 resistance genes that confer resistance to β-lactams, kanamycin, or cotrimoxazole (R* indicates presence of additional extended-spectrum β-lactamase gene; R** indicates presence of mcr-1 gene conferring additional resistance to colistin). The strain origin (country and source of isolation) is represented by flags and human, animal, or water symbols. CC, clonal complex; HPI, high-pathogenicity island (presence of chromosomal locus encoding the siderophore yersiniabactin); SNP, single-nucleotide polymorphism; ST, sequence type. Scale bar indicates nucleotide substitutions per site.