Volume 24, Number 5—May 2018
Research
Epidemic Dynamics of Vibrio parahaemolyticus Illness in a Hotspot of Disease Emergence, Galicia, Spain
Figure 2
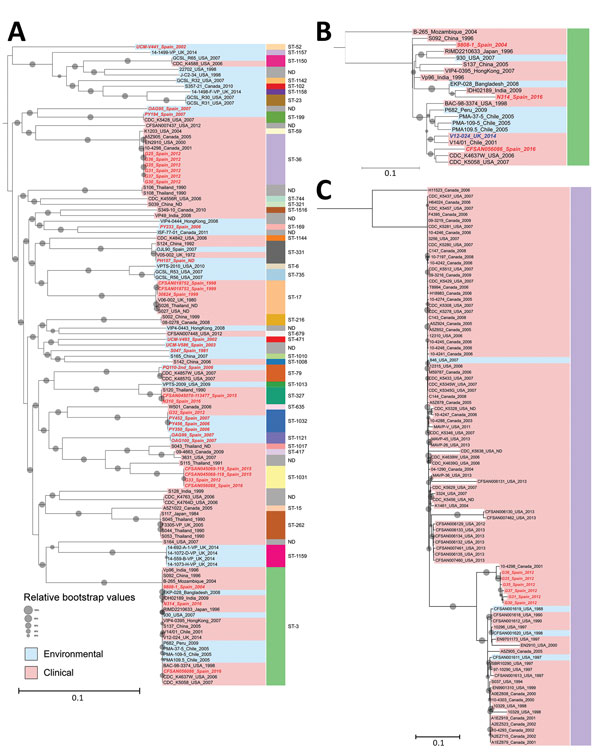
Figure 2. Phylogeny of Vibrio parahaemolyticus isolates from Galicia, Spain. A) Phylogenetic inference of the 42 genomes from Spain identified in this study (red text) along with all other genomes identified in the same clusters by the global phylogeny with their corresponding sequence types (STs). B) Phylogenetic tree of genomes belonging to ST3 (pandemic clone). C) Phylogenetic tree of genomes included in ST36 in the global phylogeny. Gray dots indicate bootstrap values supporting the nodes; dot sizes indicate 80% (smallest) to 100% (largest). Values <80% are not shown. Scale bars represent nucleotide substitutions per site. ND, not determined.
Page created: April 17, 2018
Page updated: April 17, 2018
Page reviewed: April 17, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.