Foot-and-Mouth Disease in the Middle East Caused by an A/ASIA/G-VII Virus Lineage, 2015–2016
Katarzyna Bachanek-Bankowska

, Antonello Di Nardo, Jemma Wadsworth, Elisabeth K.M. Henry, Ünal Parlak, Anna Timina, Alexey Mischenko, Ibrahim Ahmad Qasim, Darab Abdollahi, Munawar Sultana, M. Anwar Hossain, Donald P. King, and Nick J. Knowles
Author affiliations: The Pirbright Institute, Woking, UK (K. Bachanek-Bankowska, A. Di Nardo, J. Wadsworth, E.K.M. Henry, D.P. King, N.J. Knowles); Foot-and-Mouth Disease Institute, Ankara, Turkey (Ü. Parlak); Federal Centre for Animal Health, Vladimir, Russia (A. Timina, A. Mischenko); Ministry of Environment, Water and Agriculture, Riyadh, Saudi Arabia (I.A. Qasim); Iran Veterinary Organization, Tehran, Iran (D. Abdollahi); University of Dhaka, Dhaka, Bangladesh (M. Sultana, M.A. Hossain)
Main Article
Figure 2
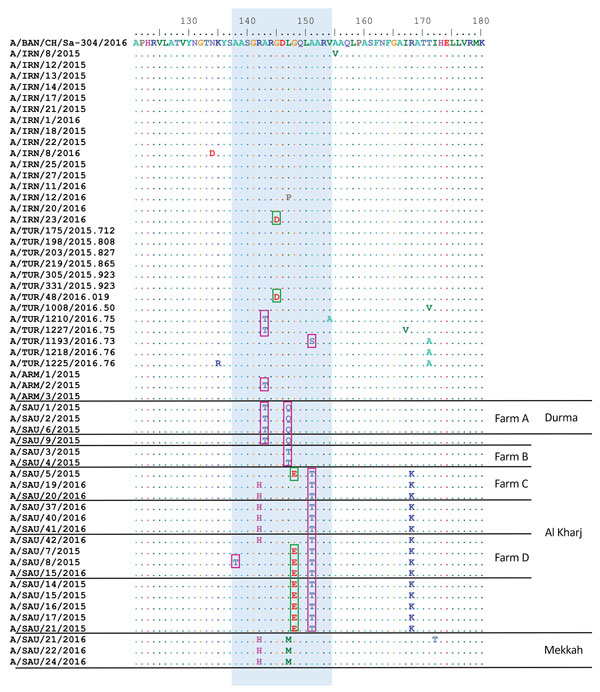
Figure 2. Comparison of predicted amino acid sequences of foot-and-mouth disease viruses showing changes in major antigenic sites. Predicted amino acid sequences for samples collected during outbreaks of foot-and-mouth disease during 2015–2016 were compared with A/BAN/CH/Sa-304/2016 virus sequence. Blue shading indicates conservative changes within antigenic site 1, pink boxes indicate hydrophobic to hydrophilic substitutions, and green boxes indicate hydrophobic to acidic substitutions. Dots indicate sequence identity. Amino acid residues are colored according to their physicochemical properties. Four large dairy farms (containing >1,000 lactating cows), which were multiply sampled, are indicated. ARM, Armenia; BAN, Bangladesh; IRN, Iran; SAU, Saudi Arabia; TUR, Turkey.
Main Article
Page created: May 17, 2018
Page updated: May 17, 2018
Page reviewed: May 17, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
