Volume 24, Number 8—August 2018
Research
Human Norovirus Replication in Human Intestinal Enteroids as Model to Evaluate Virus Inactivation
Figure 2
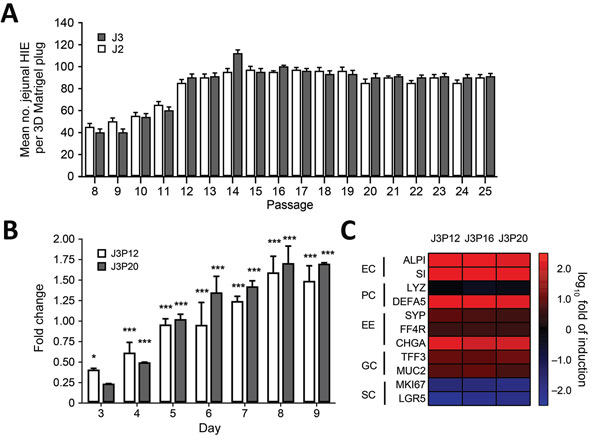
Figure 2. Characterization of differentiated and undifferentiated HIE in a study of human norovirus replication in HIEs. A) Quantification of undifferentiated HIE generated on each passage. Undifferentiated HIEs derived from 2 donors (J2P7 and J3P7) frozen at passage 7 (P7) were recovered from LN2 and embedded in Matrigel (BD Biosciences, San Jose, CA, USA) (4 plugs per HIE). Cell count was performed at day 7. On that day, undifferentiated HIEs were split 1:2 or 1:3, depending on density, and seeded again in Matrigel. All available wells (n > 4) per passage were counted. Error bars indicate SD. B) Analysis of stem cell proliferation marker gene LGR5 expression by quantitative reverse transcription PCR in undifferentiated HIEs. HIEs were embedded into Matrigel, seeded in individual wells, and cultured in the presence of complete media with growth factors. RNA was isolated from 2 wells at 1 hour postseed (day 0) and each day during days 3–9. LGR5 expression was normalized to GAPDH and expressed as fold change relative to day 0 (n = 2 wells/bar). Two different passages were assayed (P12 and P20). Error bars indicate SDs; asterisks indicate significant difference from day 0: *p<0.05; ***p<0.001. C) Heat map based on log (2–∆∆Ct) comparing gene expression levels for markers of differentiated small intestinal epithelial cells between undifferentiated and 4-day differentiated HIE monolayers. Experiments were performed with 3 independent cell passages (P12, P16, and P20). Transcripts were normalized to GAPDH levels. Shown are markers for enterocytes (EC), Paneth cells (PC), enteroendocrine cells (EE), goblet cells (GC), and stem cells (SC). Gene symbols: ALPI, intestinal-type alkaline phosphatase; CHGA, chromogranin A; DEFA5, defensin α 5; FFA4R, free fatty acid receptor 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LGR5, leucin-rich repeat-containing G-protein-couple receptor 5; LYZ, lysosyme; MKI67, marker of proliferation Ki-67; MUC2, mucin 2; SI, sucrose isomaltase; SYP, synaptophysin; TFF3, trefoil factor 3. 3D, 3-dimensional; HIE, human intestinal enteroid.