Middle East Respiratory Syndrome Coronavirus, Saudi Arabia, 2017–2018
Ahmed Hakawi, Erica Billig Rose

, Holly M. Biggs, Xiaoyan Lu, Mutaz Mohammed, Osman Abdalla, Glen R. Abedi, Ali A. Alsharef, Aref Ali Alamri, Samar Ahmad Bereagesh, Kamel M. Al Dosari, Saad Abdullah Ashehri, Waad Ghassan Fakhouri, Saleh Zaid Alzaid, Stephen Lindstrom, Susan I. Gerber, Abdullah Asiri, Hani Jokhdar, and John T. Watson
Author affiliations: Ministry of Health, Riyadh, Saudi Arabia (A. Hakawi, M. Mohammed, O. Abdalla, A.A. Alsharef, A.A. Alamri, S.A. Bereagesh, K.M. Al Dosari, S.A. Ashehri, W.G. Fakhouri, S.Z. Alzaid, A. Asiri, H. Jokhdar); Centers for Disease Control and Prevention, Atlanta, Georgia, USA (E.B. Rose, H.M. Biggs, X. Lu, G.R. Abedi, S. Lindstrom, S.I. Gerber, J.T. Watson)
Main Article
Figure
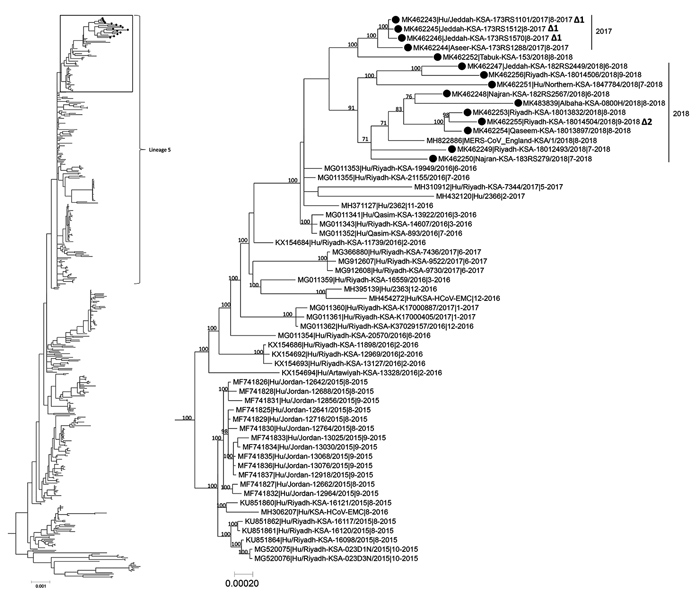
Figure. Phylogenetic tree of MERS-CoV whole-genome sequences obtained in Saudi Arabia (black dots) compared with 472 previously published human and camel genome sequences from GenBank. Tree inferred using MrBayes version 3.2.6 (https://nbisweden.github.io/MrBayes) under a general time-reversible model of nucleotide substitution with 4 categories of γ-distributed rate heterogeneity and a proportion of invariant sites. Box at the top of the tree on the left shows location of the tree on the right, showing lineage 5. Δ1 indicates 3-nt in-frame deletion in open reading frame 4a and Δ2 indicates 66-nt in-frame deletion in open reading frame 1a. Clade-credibility values >70% are indicated at selected nodes. Sequences from this study include GenBank accession numbers MK462243–MK462256 and MK483839. Scale bars indicate nucleotide substitutions per site. MERS-CoV, Middle East respiratory syndrome coronavirus; Hu, human; KSA, Kingdom of Saudi Arabia.
Main Article
Page created: October 16, 2019
Page updated: October 16, 2019
Page reviewed: October 16, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
