Volume 25, Number 12—December 2019
CME ACTIVITY - Research
Streptococcus suis–Associated Meningitis, Bali, Indonesia, 2014–2017
Figure 2
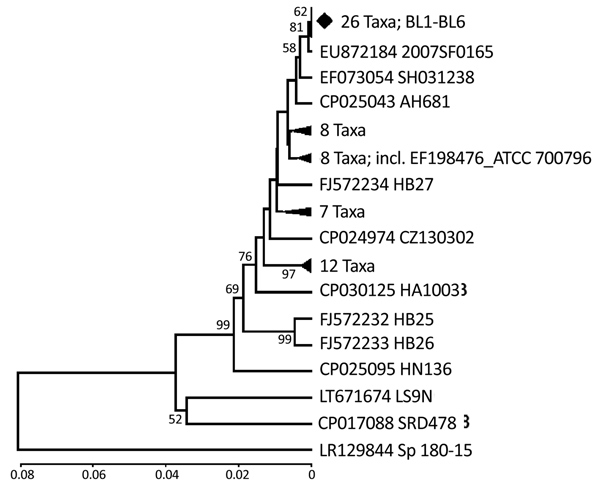
Figure 2. Phylogenetic relationships of the glutamate dehydrogenase gene fragment of Streptococcus suis isolated from humans in Denpasar, Bali, Indonesia (BL1-BL6 taxa), with sequences data of S. suis available in GenBank. The phylogeny was inferred using unweighted pair group method with arithmetic mean (13). The GenBank accession number and strain name are written as taxon name. To minimize crowding, some tree branches were condensed. The number of taxa in each condensed branch is indicated. The location of standard American Type Culture Collection isolate (GenBank accession no. EF198476) is shown. Respective gene sequence of full-genome data of S. pneumoniae (accession no. LR129844) was co-analyzed as outgroup. The percentage of replicate trees in which the associated taxa clustered in the bootstrap test (1,000 replicates) are shown next to the branches (16). Bootstrap values of <50% are not shown. The genetic distances were computed using the Kimura 2-parameter method (17). Phylogenetic analyses were conducted in MEGA6 (18). Scale bar indicates nucleotide substitutions per site.
References
- van de Beek D, Brouwer M, Hasbun R, Koedel U, Whitney CG, Wijdicks E. Community-acquired bacterial meningitis. Nat Rev Dis Primers. 2016;2:16074. DOIPubMedGoogle Scholar
- van Samkar A, Brouwer MC, Schultsz C, van der Ende A, van de Beek D. Streptococcus suis meningitis: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2015;9:
e0004191 . DOIPubMedGoogle Scholar - Feng Y, Zhang H, Wu Z, Wang S, Cao M, Hu D, et al. Streptococcus suis infection: an emerging/reemerging challenge of bacterial infectious diseases? Virulence. 2014;5:477–97. DOIPubMedGoogle Scholar
- van Samkar A, Brouwer MC, van der Ende A, van de Beek D. Zoonotic bacterial meningitis in human adults. Neurology. 2016;87:1171–9. DOIPubMedGoogle Scholar
- Gottschalk M, Fittipaldi N, Segura M. Streptococcus suis meningitis. In: Christodoulides M, editor. Meningitis: cellular and molecular basis. Oxford (UK): CAB International; 2013. p. 184–98.
- Wertheim HF, Nghia HD, Taylor W, Schultsz C. Streptococcus suis: an emerging human pathogen. Clin Infect Dis. 2009;48:617–25. DOIPubMedGoogle Scholar
- Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect. 2014;3:
e45 . DOIPubMedGoogle Scholar - Scarborough M, Thwaites GE. The diagnosis and management of acute bacterial meningitis in resource-poor settings. Lancet Neurol. 2008;7:637–48. DOIPubMedGoogle Scholar
- Lehman DC, Mahon CR, Suvarna K. Streptococcus, Enterococcus, and other catalase-negative, gram-positive cocci. In: Mahon CR, Lehman DC, Manuselis G, editors. Textbook of diagnostic microbiology. 5th ed. Maryland Heights (MO): Saunders Elsivier; 2016. p. 328–48.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement (M100-S27). Wayne (PA): The Institute; 2017. p. 250.
- Ishida S, Tien HT, Osawa R, Tohya M, Nomoto R, Kawamura Y, et al. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J Microbiol Methods. 2014;107:66–70. DOIPubMedGoogle Scholar
- Okwumabua O, O’Connor M, Shull E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett. 2003;218:79–84. DOIPubMedGoogle Scholar
- Sneath PHA, Sokal RR. Numerical taxonomy. San Francisco: Freeman; 1973.
- Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, et al. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One. 2013;8:
e72070 . DOIPubMedGoogle Scholar - Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. DOIPubMedGoogle Scholar
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. DOIPubMedGoogle Scholar
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Tien HT, Nishibori T, Nishitani Y, Nomoto R, Osawa R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet Microbiol. 2013;162:842–9. DOIPubMedGoogle Scholar
- Southeast Asia Infectious Disease Clinical Research Network. Causes and outcomes of sepsis in southeast Asia: a multinational multicentre cross-sectional study. Lancet Glob Health. 2017;5:e157–67. DOIPubMedGoogle Scholar
- Nugroho W, Cargill CF, Putra IM, Kirkwood RN, Trott DJ, Salasia SI, et al. Investigations of selected pathogens among village pigs in Central Papua, Indonesia. Trop Anim Health Prod. 2016;48:29–36. DOIPubMedGoogle Scholar
- Zalas-Wiecek P, Michalska A, Grabczewska E, Olczak A, Pawlowska M, Gospodarek E. Human meningitis caused by Streptococcus suis. J Med Microbiol. 2013;62:483–5. DOIPubMedGoogle Scholar
- Ngo TH, Tran TB, Tran TT, Nguyen VD, Campbell J, Pham HA, et al. Slaughterhouse pigs are a major reservoir of Streptococcus suis serotype 2 capable of causing human infection in southern Vietnam. PLoS One. 2011;6:
e17943 . DOIPubMedGoogle Scholar - Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, et al.; Streptococcus suis study groups. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:914–20. DOIPubMedGoogle Scholar
- Nghia HD, Tu TP, Wolbers M, Thai CQ, Hoang NV, Nga TV, et al. Risk factors of Streptococcus suis infection in Vietnam. A case-control study. Erratum in: PLoS One. 2011;6(4); 2012;7(5). PLoS One. 2011;6:e17604.
- Takeuchi D, Kerdsin A, Akeda Y, Chiranairadul P, Loetthong P, Tanburawong N, et al. Impact of food safety campaign on Streptococcus suis infection in humans in Thailand. Am J Trop Med Hyg. 2017;96:1370–7. DOIPubMedGoogle Scholar
- Navacharoen N, Chantharochavong V, Hanprasertpong C, Kangsanarak J, Lekagul S. Hearing and vestibular loss in Streptococcus suis infection from swine and traditional raw pork exposure in northern Thailand. J Laryngol Otol. 2009;123:857–62. DOIPubMedGoogle Scholar
- Takeuchi D, Kerdsin A, Pienpringam A, Loetthong P, Samerchea S, Luangsuk P, et al. Population-based study of Streptococcus suis infection in humans in Phayao Province in northern Thailand. PLoS One. 2012;7:
e31265 . DOIPubMedGoogle Scholar - Huong VT, Ha N, Huy NT, Horby P, Nghia HD, Thiem VD, et al. Epidemiology, clinical manifestations, and outcomes of Streptococcus suis infection in humans. Emerg Infect Dis. 2014;20:1105–14. DOIPubMedGoogle Scholar
- Teekakirikul P, Wiwanitkit V. Streptococcus suis infection: overview of case reports in Thailand. Southeast Asian J Trop Med Public Health. 2003;34(Suppl 2):178–83.PubMedGoogle Scholar
- Heidt MC, Mohamed W, Hain T, Vogt PR, Chakraborty T, Domann E. Human infective endocarditis caused by Streptococcus suis serotype 2. J Clin Microbiol. 2005;43:4898–901. DOIPubMedGoogle Scholar
- van de Beek D, de Gans J, Spanjaard L, Weisfelt M, Reitsma JB, Vermeulen M. Clinical features and prognostic factors in adults with bacterial meningitis. N Engl J Med. 2004;351:1849–59. DOIPubMedGoogle Scholar
- Fillo S, Mancini F, Anselmo A, Fortunato A, Rezza G, Lista F, et al. Draft genome sequence of Streptococcus suis strain SsRC-1, a human isolate from a fatal case of toxic shock syndrome. Genome Announc. 2018;6:e00447–18. DOIPubMedGoogle Scholar
- Wangsomboonsiri W, Luksananun T, Saksornchai S, Ketwong K, Sungkanuparph S. Streptococcus suis infection and risk factors for mortality. J Infect. 2008;57:392–6. DOIPubMedGoogle Scholar
- Robertson ID, Blackmore DK. Experimental studies on the comparative infectivity and pathogenicity of Streptococcus suis type 2. I. Porcine and human isolates in pigs. Epidemiol Infect. 1990;105:469–78. DOIPubMedGoogle Scholar
- Taniyama D, Sakurai M, Sakai T, Kikuchi T, Takahashi T. Human case of bacteremia due to Streptococcus suis serotype 5 in Japan: The first report and literature review. IDCases. 2016;6:36–8. DOIPubMedGoogle Scholar
- Kerdsin A, Hatrongjit R, Gottschalk M, Takeuchi D, Hamada S, Akeda Y, et al. Emergence of Streptococcus suis serotype 9 infection in humans. J Microbiol Immunol Infect. 2017;50:545–6. DOIPubMedGoogle Scholar
- Nghia HD, Hoa NT, Linh D, Campbell J, Diep TS, Chau NV, et al. Human case of Streptococcus suis serotype 16 infection. Emerg Infect Dis. 2008;14:155–7. DOIPubMedGoogle Scholar
- Callejo R, Prieto M, Salamone F, Auger JP, Goyette-Desjardins G, Gottschalk M. Atypical Streptococcus suis in man, Argentina, 2013. Emerg Infect Dis. 2014;20:500–2. DOIPubMedGoogle Scholar
- Kerdsin A, Gottschalk M, Hatrongjit R, Hamada S, Akeda Y, Oishi K. Fatal septic meningitis in child caused by Streptococcus suis serotype 24. Emerg Infect Dis. 2016;22:1519–20. DOIPubMedGoogle Scholar
- Hatrongjit R, Kerdsin A, Gottschalk M, Takeuchi D, Hamada S, Oishi K, et al. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect Dis. 2015;15:392. DOIPubMedGoogle Scholar
- Lun ZR, Wang QP, Chen XG, Li AX, Zhu XQ. Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect Dis. 2007;7:201–9. DOIPubMedGoogle Scholar
- Fongcom A, Pruksakorn S, Netsirisawan P, Pongprasert R, Onsibud P. Streptococcus suis infection: a prospective study in northern Thailand. Southeast Asian J Trop Med Public Health. 2009;40:511–7.PubMedGoogle Scholar
- Tsai HY, Liao CH, Liu CY, Huang YT, Teng LJ, Hsueh PR. Streptococcus suis infection in Taiwan, 2000-2011. Diagn Microbiol Infect Dis. 2012;74:75–7. DOIPubMedGoogle Scholar
- Gomez-Torres J, Nimir A, Cluett J, Aggarwal A, Elsayed S, Soares D, et al. Human case of Streptococcus suis disease, Ontario, Canada. Emerg Infect Dis. 2017;23:2107–9. DOIPubMedGoogle Scholar