Volume 25, Number 12—December 2019
Dispatch
West Nile Virus in Wildlife and Nonequine Domestic Animals, South Africa, 2010–2018
Figure 2
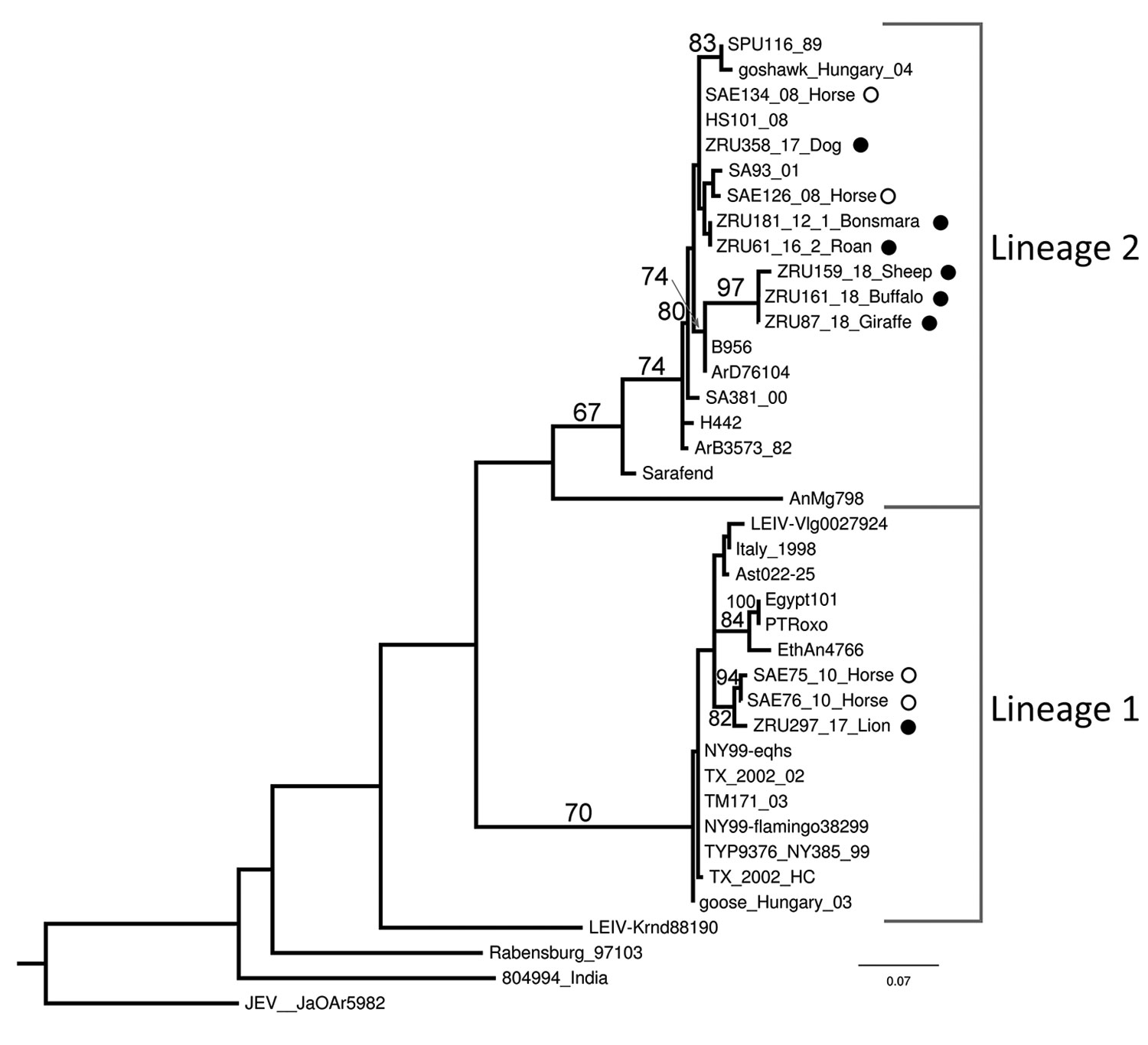
Figure 2. Maximum-likelihood phylogram of the partial (215-nt) nonstructural protein gene used for identification of West Nile virus infection in wildlife and nonequine domestic animals, South Africa, 2010–2018. Tree was generated with RAXML (https://cme.h-its.org/exelixis/web/software/raxml) using the general time-reversible plus gamma model with 39 taxa and the AutoMRE bootstopping function invoked (bootstraps >65 as branch support). Black circles indicate wildlife and nonequine domestic animal sequences from this study; open circles indicate horse sequences (3,12). Reference strains, GenBank accession numbers, and origins are as indicated in (4). GenBank accession numbers for the newly sequenced strains are ZRU87_18, MN270988; ZRU159_18_SA, MN270989; ZRU161_18_SA, MN27099; and ZRU181_12_1, KY176733. The sequences for strains ZRU358/17, ZRU061/16/2, and ZRU297/17 were <200 bp long and therefore could not be submitted to GenBank; the sequence data are available from the authors. Scale bar indicates nucleotide substitutions per site.
References
- Murray KO, Walker C, Gould E. The virology, epidemiology, and clinical impact of West Nile virus: a decade of advancements in research since its introduction into the Western Hemisphere. Epidemiol Infect. 2011;139:807–17. DOIPubMedGoogle Scholar
- Pauli G, Bauerfeind U, Blümel J, Burger R, Drosten C, Gröner A, et al. West nile virus. Transfus Med Hemother. 2013;40:265–84.PubMedGoogle Scholar
- Venter M, Pretorius M, Fuller JA, Botha E, Rakgotho M, Stivaktas V, et al. West Nile virus lineage 2 in horses and other animals with neurologic disease, South Africa, 2008–2015. Emerg Infect Dis. 2017;23:2060–4. DOIPubMedGoogle Scholar
- Gould LH, Fikrig E. West Nile virus: a growing concern? J Clin Invest. 2004;113:1102–7. DOIPubMedGoogle Scholar
- Zaayman D, Human S, Venter M. A highly sensitive method for the detection and genotyping of West Nile virus by real-time PCR. J Virol Methods. 2009;157:155–60. DOIPubMedGoogle Scholar
- O’Dell N, Arnot L, Janisch CE, Steyl JC. Clinical presentation and pathology of suspected vector transmitted African horse sickness in South African domestic dogs from 2006 to 2017. Vet Rec. 2018;182:715. DOIPubMedGoogle Scholar
- Blackburn NK, Reyers F, Berry WL, Shepherd AJ. Susceptibility of dogs to West Nile virus: a survey and pathogenicity trial. J Comp Pathol. 1989;100:59–66. DOIPubMedGoogle Scholar
- van Niekerk S, Human S, Williams J, van Wilpe E, Pretorius M, Swanepoel R, et al. Sindbis and Middelburg Old World alphaviruses associated with neurologic disease in horses, South Africa. Emerg Infect Dis. 2015;21:2225–9. DOIPubMedGoogle Scholar
- Van Eeden C, Zaayman D, Venter M. A sensitive nested real-time RT-PCR for the detection of Shuni virus. J Virol Methods. 2014;195:100–5. DOIPubMedGoogle Scholar
- van Niekerk M, Freeman M, Paweska JT, Howell PG, Guthrie AJ, Potgieter AC, et al. Variation in the NS3 gene and protein in South African isolates of bluetongue and equine encephalosis viruses. J Gen Virol. 2003;84:581–90. DOIPubMedGoogle Scholar
- Yaeger M, Yoon K-J, Schwartz K, Berkland L. West Nile virus meningoencephalitis in a Suri alpaca and Suffolk ewe. J Vet Diagn Invest. 2004;16:64–6. DOIPubMedGoogle Scholar
- Venter M, Human S, van Niekerk S, Williams J, van Eeden C, Freeman F. Fatal neurologic disease and abortion in mare infected with lineage 1 West Nile virus, South Africa. Emerg Infect Dis. 2011;17:1534–6. DOIPubMedGoogle Scholar
- Blitvich BJ, Marlenee NL, Hall RA, Calisher CH, Bowen RA, Roehrig JT, et al. Epitope-blocking enzyme-linked immunosorbent assays for the detection of serum antibodies to west nile virus in multiple avian species. J Clin Microbiol. 2003;41:1041–7. DOIPubMedGoogle Scholar
- Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–8. DOIPubMedGoogle Scholar
- Zaayman D, Venter M. West Nile virus neurologic disease in humans, South Africa, September 2008-may 2009. Emerg Infect Dis. 2012;18:2051–4. DOIPubMedGoogle Scholar