Volume 25, Number 3—March 2019
Research Letter
Detection of Influenza C Virus Infection among Hospitalized Patients, Cameroon
Figure
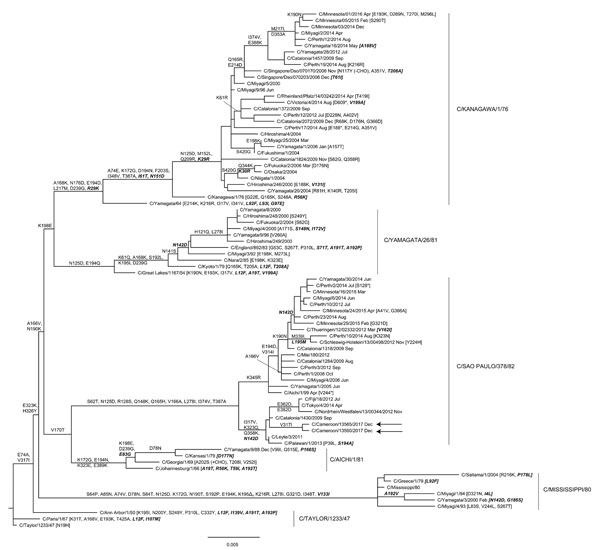
Figure. Hemagglutinin-esterase gene phylogeny for influenza C viruses detected in Cameroon compared with reference viruses. The phylogeny is based on 80 full-length open-reading frames downloaded from the GISAID EpiFlu database (https://www.gisaid.org), with signal peptide coding regions and stop codons removed, yielding products of 1,923 nt. The phylogeny was estimated by using RaxML version 8.2.X (https://sco.h-its.org/exelixis/software.html) with a general time-reversible plus gamma substitution model and then annotated with amino acid substitutions defining nodes and individual virus gene products by using treesub (https://github.com/tamuri/treesub/blob/master/README.md). The phylogeny was visualized by using FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree). Sequences for the 2 viruses from Cameroon (arrows) have been deposited in the EpiFlu database under accession nos. EPI1259829 (C/Cameroon/13560/2017) and EPI1259835 (C/Cameroon/13565/2017). Scale bar indicates nucleotide substitutions per site.