Volume 25, Number 9—September 2019
Research Letter
Fatal Case of Lassa Fever, Bangolo District, Côte d’Ivoire, 2015
Figure
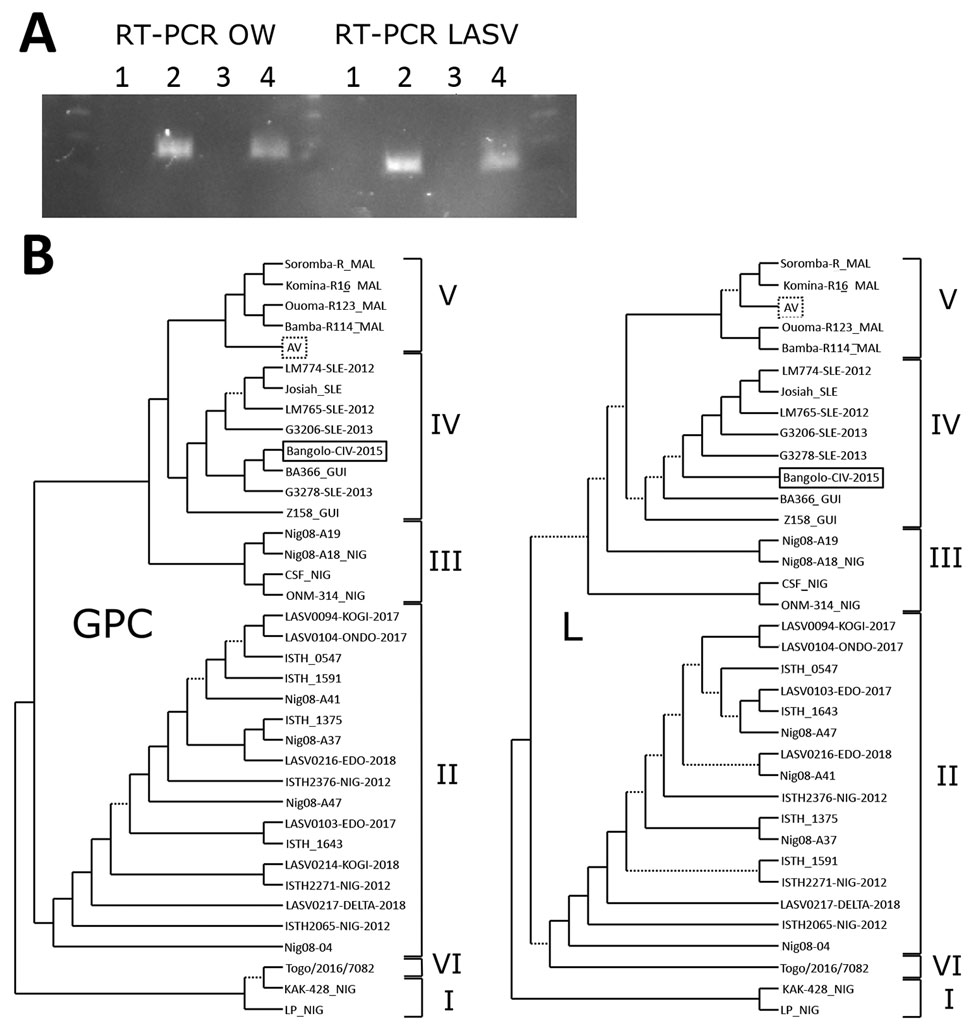
Figure. Analysis of LASV strains, Bangolo District, Côte d’Ivoire, 2015. A) RT-PCR analysis of human serum samples 132/16 and 001/15 by using OW and LASV RT-PCRs. Lane 1, negative control; lane 2, positive control; lane 3, 132/16; lane 4, 001/15. B) Phylogenetic analysis of LASV strains. Trees were inferred by using the PhyML Smart Model Selection (5) general time-reversible plus gamma plus proportion of invariable sites model with 200 bootstrap replicates. Poorly supported branches with bootstrap values <0.50 are indicated by dotted lines. Lassa virus lineages are indicated by the Roman numerals on the right. The Bangolo-CIV-2015 strain (solid box), which was isolated in this study, appears to be related to clade IV, and the AV strain (dotted box) is related to clade V. GPC, glycoprotein complex gene; L, large RNA segment; LASV, Lassa virus; OW, Old World; RT-PCR, reverse transcription PCR.
References
- Günther S, Emmerich P, Laue T, Kühle O, Asper M, Jung A, et al. Imported lassa fever in Germany: molecular characterization of a new lassa virus strain. Emerg Infect Dis. 2000;6:466–76. DOIPubMedGoogle Scholar
- Kouadio L, Nowak K, Akoua-Koffi C, Weiss S, Allali BK, Witkowski PT, et al. Lassa virus in multimammate rats, Côte d’Ivoire, 2013. Emerg Infect Dis. 2015;21:1481–3. DOIPubMedGoogle Scholar
- Akoua-Koffi C, Ter Meulen J, Legros D, Akran V, Aïdara M, Nahounou N, et al. Detection of anti-Lassa antibodies in the Western Forest area of the Ivory Coast [in French]. Med Trop (Mars). 2006;66:465–8.
- Andersen KG, Shapiro BJ, Matranga CB, Sealfon R, Lin AE, Moses LM, et al.; Viral Hemorrhagic Fever Consortium. Clinical sequencing uncovers origins and evolution of Lassa virus. Cell. 2015;162:738–50. DOIPubMedGoogle Scholar
- Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34:2422–4. DOIPubMedGoogle Scholar
- Lecompte E, Fichet-Calvet E, Daffis S, Koulémou K, Sylla O, Kourouma F, et al. Mastomys natalensis and Lassa fever, West Africa. Emerg Infect Dis. 2006;12:1971–4. DOIPubMedGoogle Scholar
- Kofman A, Choi MJ, Rollin PE. Lassa fever in travelers from West Africa, 1969–2016. Emerg Infect Dis. 2019;25:245–8. DOIPubMedGoogle Scholar
- Kafetzopoulou LE, Pullan ST, Lemey P, Suchard MA, Ehichioya DU, Pahlmann M, et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science. 2019;363:74–7. DOIPubMedGoogle Scholar
- Coulibaly-N’Golo D, Allali B, Kouassi SK, Fichet-Calvet E, Becker-Ziaja B, Rieger T, et al. Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Côte d’Ivoire: implications for evolution of arenaviruses in Africa. PLoS One. 2011;6:
e20893 . DOIPubMedGoogle Scholar