Candida auris in Germany and Previous Exposure to Foreign Healthcare
Axel Hamprecht, Amelia E. Barber, Sibylle C. Mellinghoff, Philipp Thelen, Grit Walther, Yanying Yu, Priya Neurgaonkar, Thomas Dandekar, Oliver A. Cornely, Ronny Martin, Oliver Kurzai

, on behalf of the German Candida auris Study Group
Author affiliations: German Centre for Infection Research, Cologne, Germany (A. Hamprecht, S.C. Mellinghoff, O.A. Cornely); University of Cologne, Cologne (A. Hamprecht, O.A. Cornely); Leibniz Institute for Natural Product Research and Infection Biology–Hans-Knoell-Institute, Jena, Germany (A.E. Barber, G. Walther, O. Kurzai); University Hospital Cologne, Cologne (S.C. Mellinghoff, P. Thelen); University of Würzburg, Würzburg, Germany (Y. Yu, P. Neurgaonkar, T. Dandekar, R. Martin, O. Kurzai)
Main Article
Figure
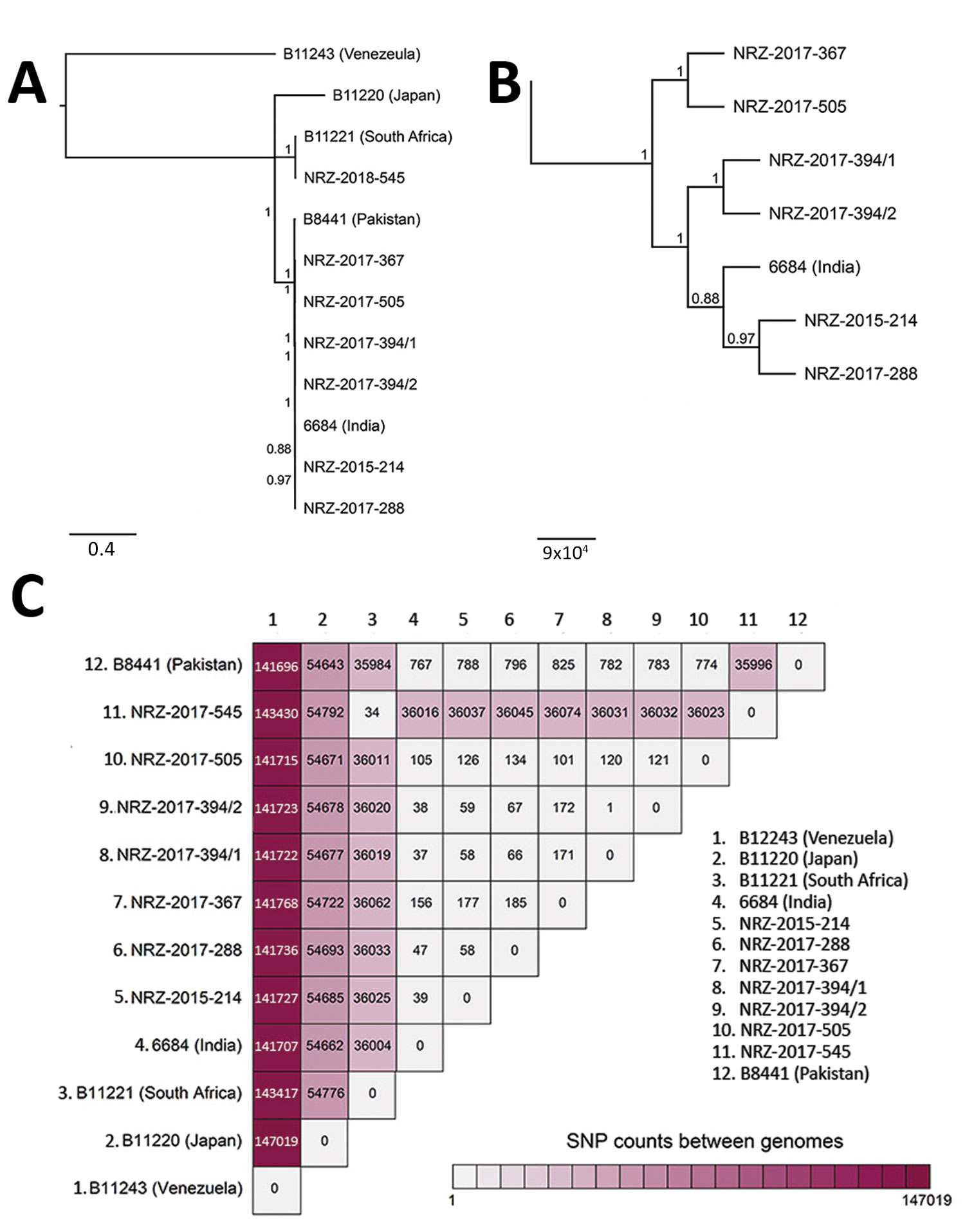
Figure. Genetic relationships of Candida auris isolates based on whole-genome sequencing SNP analysis. A) Maximum-likelihood phylogeny of C. auris isolates from Germany (indicated by NRZ prefix) inferred to reveal a possible geographic origin. The isolates were contrasted against strains representing the 4 different clades of C. auris: South American (strain B114243 from Venezuela), East Asian (B11220 from Japan), South African (B11221 from South Africa), and South Asian (B8441 from Pakistan and 6684 from India). B) Higher resolution of the tree shown in panel A to better visualize the relationship between the isolates belonging to the South Asian clade. Scale bars in panels A and B indicate nucleotide substitutions per site. C) SNP counts between the genomes of the isolates from Germany and the representative strains from the different clades. SNP, single-nucleotide polymorphism.
Main Article
Page created: August 21, 2019
Page updated: August 21, 2019
Page reviewed: August 21, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
