Volume 26, Number 11—November 2020
Research
Streptococcus pneumoniae Serotype 12F-CC4846 and Invasive Pneumococcal Disease after Introduction of 13-Valent Pneumococcal Conjugate Vaccine, Japan, 2015–2017
Figure 1
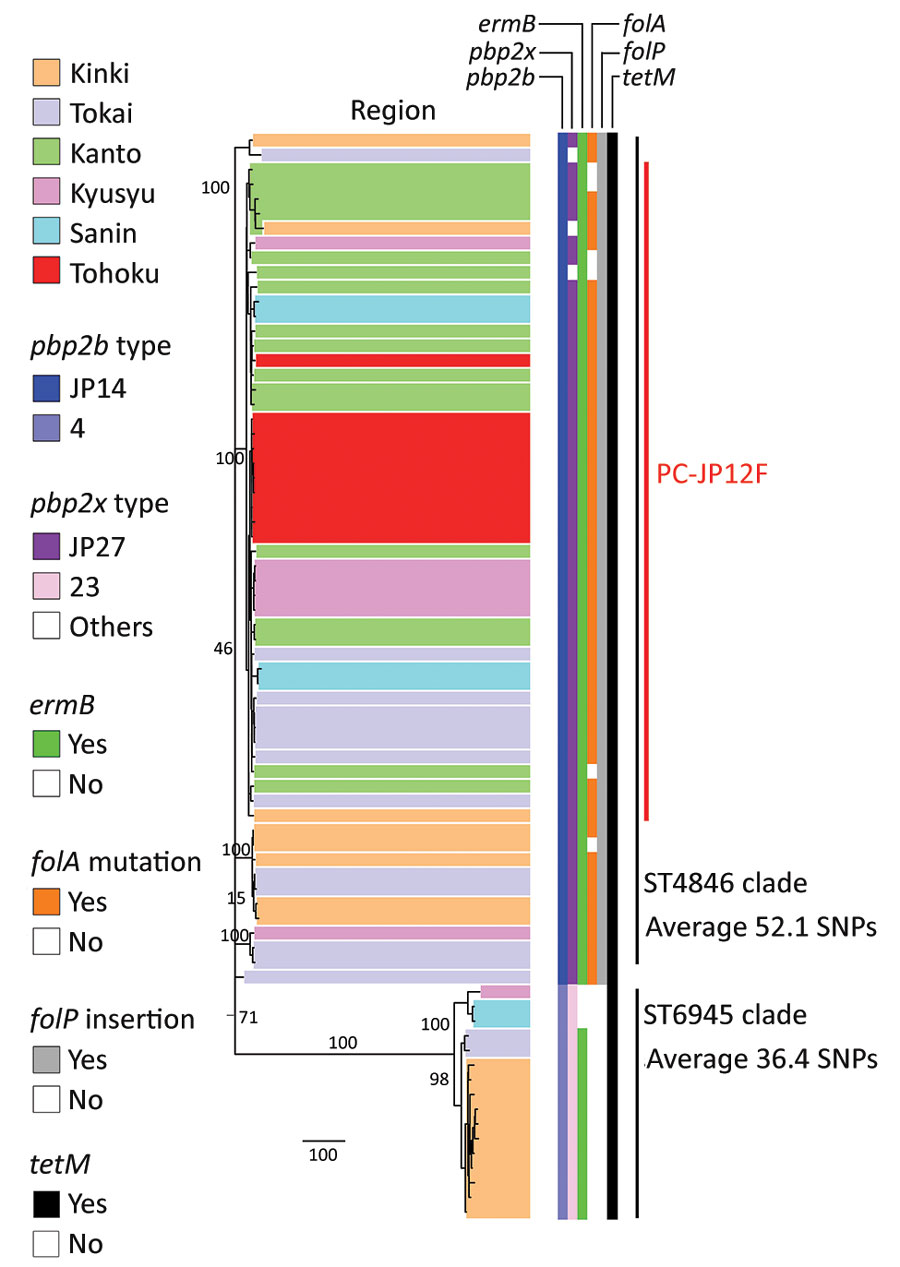
Figure 1. Recombination-free maximum-likelihood tree of Streptococcus pneumoniae serotype 12F-CC4846 isolates in Japan, created by using Gubbins (27). Two clusters were generated: 1 comprised only sequence type (ST) 4846 isolates and the other comprised only ST6945 isolates. All isolates had pbp1a-13. The pbp2x type “others” included pbp2x-JP23, pbp2x-JP58, and pbp2x-JP59. The geographic locations of the described regions in this figure are shown in the Appendix. The numbers on the branches indicate bootstrap values. SNP, single-nucleotide polymorphism; ST, sequence type.
References
- Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-15. Lancet Glob Health. 2018;6:e744–57. DOIPubMedGoogle Scholar
- Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, et al. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev. 2015;28:871–99. DOIPubMedGoogle Scholar
- Waight PA, Andrews NJ, Ladhani NJ, Sheppard CL, Slack MP, Miller E. Effect of the 13-valent pneumococcal conjugate vaccine on invasive pneumococcal disease in England and Wales 4 years after its introduction: an observational cohort study. Lancet Infect Dis. 2015;15:629. DOIPubMedGoogle Scholar
- Abat C, Raoult D, Rolain JM. Dramatic decrease of Streptococcus pneumoniae infections in Marseille, 2003-2014. Eur J Clin Microbiol Infect Dis. 2015;34:2081–7. DOIPubMedGoogle Scholar
- Camilli R, D’Ambrosio F, Del Grosso M, Pimentel de Araujo F, Caporali MG, Del Manso M, et al.; Pneumococcal Surveillance Group. Impact of pneumococcal conjugate vaccine (PCV7 and PCV13) on pneumococcal invasive diseases in Italian children and insight into evolution of pneumococcal population structure. Vaccine. 2017;35(35 Pt B):4587–93. DOIPubMedGoogle Scholar
- Moore MR, Link-Gelles R, Schaffner W, Lynfield R, Holtzman C, Harrison LH, et al. Effectiveness of 13-valent pneumococcal conjugate vaccine for prevention of invasive pneumococcal disease in children in the USA: a matched case-control study. Lancet Respir Med. 2016;4:399–406. DOIPubMedGoogle Scholar
- Chiba N, Morozumi M, Shouji M, Wajima T, Iwata S, Ubukata K; Invasive Pneumococcal Diseases Surveillance Study Group. Changes in capsule and drug resistance of Pneumococci after introduction of PCV7, Japan, 2010-2013. Emerg Infect Dis. 2014;20:1132–9. DOIPubMedGoogle Scholar
- Nakano S, Fujisawa T, Ito Y, Chang B, Suga S, Noguchi T, et al. Serotypes, antimicrobial susceptibility, and molecular epidemiology of invasive and non-invasive Streptococcus pneumoniae isolates in paediatric patients after the introduction of 13-valent conjugate vaccine in a nationwide surveillance study conducted in Japan in 2012-2014. Vaccine. 2016;34:67–76. DOIPubMedGoogle Scholar
- Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, et al. Nationwide surveillance of paediatric invasive and non-invasive pneumococcal disease in Japan after the introduction of the 13-valent conjugated vaccine, 2015-2017. Vaccine. 2020;38:1818–24. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI document M100–S25. Wayne (PA): The Institute; 2015.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. DOIPubMedGoogle Scholar
- Chang B, Morita M, Lee KI, Ohnishi M. Complete genome sequence of a sequence type 4846 Streptococcus pneumoniae serotype 12F strain isolated from a meningitis case in Japan. Microbiol Resour Announc. 2019;8:e01632–18. DOIPubMedGoogle Scholar
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. DOIPubMedGoogle Scholar
- Metcalf BJ, Gertz RE Jr, Gladstone RA, Walker H, Sherwood LK, Jackson D, et al. Active Bacterial Core surveillance team. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect. 2016;22:60.e9–29. DOIGoogle Scholar
- Metcalf BJ, Chochua S, Gertz RE Jr, Li Z, Walker H, Tran T, et al. Active Bacterial Core surveillance team. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect. 2016;22:1002.e1–8. DOIGoogle Scholar
- Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE Jr, Walker H, et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae. MBio. 2016;7:e00756–16. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Minimum inhibitory concentrations predicted by the penicillin binding protein [cited 2020 Aug 17]. https://www.cdc.gov/streplab/pneumococcus/mic.html
- Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, et al. Spread of meropenem-resistant Streptococcus pneumoniae serotype 15A-ST63 clone in Japan, 2012–2014. Emerg Infect Dis. 2018;24:275–83. DOIPubMedGoogle Scholar
- Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, et al. Whole-genome sequencing analysis of multidrug-resistant serotype 15A Streptococcus pneumoniae in Japan and the emergence of a highly resistant serotype 15A-ST9084 clone. Antimicrob Agents Chemother. 2019;63:e02579–18. DOIPubMedGoogle Scholar
- Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, et al. Penicillin-binding protein typing, antibiotic resistance gene identification, and molecular phylogenetic analysis of meropenem-resistant Streptococcus pneumoniae serotype 19A-CC3111 strains in Japan. Antimicrob Agents Chemother. 2019;63:e00711–9. DOIPubMedGoogle Scholar
- Gladstone RA, Lo SW, Lees JA, Croucher NJ, van Tonder AJ, Corander J, et al.; Global Pneumococcal Sequencing Consortium. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine. 2019;43:338–46. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–6. DOIPubMedGoogle Scholar
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–5. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:238. DOIPubMedGoogle Scholar
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. DOIPubMedGoogle Scholar
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5:
e1000520 . DOIPubMedGoogle Scholar - Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol. 2016;33:2167–9. DOIPubMedGoogle Scholar
- Brenciani A, Bacciaglia A, Vecchi M, Vitali LA, Varaldo PE, Giovanetti E. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob Agents Chemother. 2007;51:1209–16. DOIPubMedGoogle Scholar
- Schroeder MR, Stephens DS. Macrolide resistance in Streptococcus pneumoniae. Front Cell Infect Microbiol. 2016;6:98. DOIPubMedGoogle Scholar
- Adam DC, Scotch M, MacIntyre CR. Bayesian phylogeography and pathogenic characterization of smallpox based on HA, ATI, and CrmB genes. Mol Biol Evol. 2018;35:2607–17. DOIPubMedGoogle Scholar
- Ohkusu M, Takeuchi N, Ishiwada N, Ohkusu K. Clonal spread of serotype 12F ST4846 Streptococcus pneumoniae. J Med Microbiol. 2019;68:1383–90. DOIPubMedGoogle Scholar
- Ikuse T, Habuka R, Wakamatsu Y, Nakajima T, Saitoh N, Yoshida H, et al. Local outbreak of Streptococcus pneumoniae serotype 12F caused high morbidity and mortality among children and adults. Epidemiol Infect. 2018;146:1793–6. DOIPubMedGoogle Scholar
- Nakanishi N, Yonezawa T, Tanaka S, Shirouzu Y, Naito Y, Ozaki A, et al. Assessment of the local clonal spread of Streptococcus pneumoniae serotype 12F caused invasive pneumococcal diseases among children and adults. J Infect Public Health. 2019;12:867–72. DOIPubMedGoogle Scholar
- Shimbashi R, Chang B, Tanabe Y, Takeda H, Watanabe H, Kubota T, et al.; Adult IPD Study Group. Epidemiological and clinical features of invasive pneumococcal disease caused by serotype 12F in adults, Japan. PLoS One. 2019;14:
e0212418 . DOIPubMedGoogle Scholar - Linkevicius M, Cristea V, Siira L, Mäkelä H, Toropainen M, Pitkäpaasi M, et al. Outbreak of invasive pneumococcal disease among shipyard workers, Turku, Finland, May to November 2019. Euro Surveill. 2019;24:
1900681 . DOIPubMedGoogle Scholar - González-Díaz A, Càmara J, Ercibengoa M, Cercenado E, Larrosa N, Quesada MD, et al. Emerging non-13-valent pneumococcal conjugate vaccine (PCV13) serotypes causing adult invasive pneumococcal disease in the late-PCV13 period in Spain. Clin Microbiol Infect. 2020;26:753–9. DOIPubMedGoogle Scholar
- Golden AR, Baxter MR, Davidson RJ, Martin I, Demczuk W, Mulvey MR, et al. Comparison of antimicrobial resistance patterns in Streptococcus pneumoniae from respiratory and blood cultures in Canadian hospitals from 2007–16. J Antimicrob Chemother. 2019;74(Suppl_4):iv39–iv47. DOIGoogle Scholar
- Valdarchi C, Dorrucci M, Mancini F, Farchi F, Pimentel de Araujo F, Corongiu M, et al.; FIMMG Group. Pneumococcal carriage among adults aged 50 years and older with co-morbidities attending medical practices in Rome, Italy. Vaccine. 2019;37:5096–103. DOIPubMedGoogle Scholar
- Deng X, Peirano G, Schillberg E, Mazzulli T, Gray-Owen SD, Wylie JL, et al. Whole-genome sequencing reveals the origin and rapid evolution of an emerging outbreak strain of Streptococcus pneumoniae 12F. Clin Infect Dis. 2016;62:1126–32. DOIPubMedGoogle Scholar
- Balsells E, Dagan R, Yildirim I, Gounder PP, Steens A, Muñoz-Almagro C, et al. The relative invasive disease potential of Streptococcus pneumoniae among children after PCV introduction: A systematic review and meta-analysis. J Infect. 2018;77:368–78. DOIPubMedGoogle Scholar
Page created: August 20, 2020
Page updated: October 17, 2020
Page reviewed: October 17, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.