Volume 26, Number 12—December 2020
Synopsis
Mycoplasma bovis Infections in Free-Ranging Pronghorn, Wyoming, USA
Figure 5
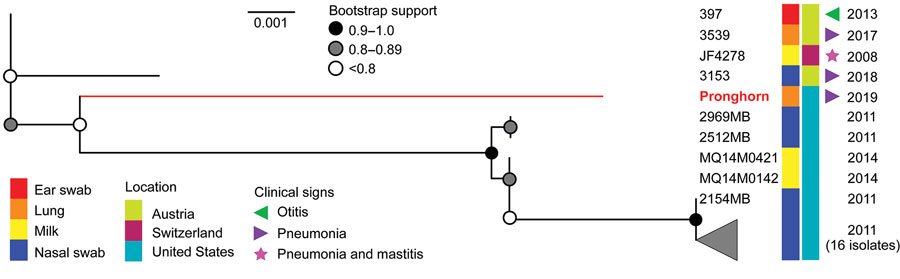
Figure 5. Phylogeny of Mycoplasma bovis isolates from free-ranging pronghorn (red branch), Wyoming, USA, February–April 2019. Pronghorn were found to be divergent from all bovine isolates with a deletion of adh-1 gene but are most similar to those recovered from cattle in the United States. This unrooted maximum-likelihood tree (10,000 bootstrap replicates) comprises all available nontypeable isolates and is based on 6 of 7 sequence typing loci. The health status of cattle sampled during 2011–2014 is unknown, and the absence of reported clinical signs does not necessarily equate to absence of disease. Scale bar indicates substitutions per site.
Page created: September 30, 2020
Page updated: November 19, 2020
Page reviewed: November 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.