Volume 26, Number 12—December 2020
Research Letter
SARS-CoV-2 in Quarantined Domestic Cats from COVID-19 Households or Close Contacts, Hong Kong, China
Figure
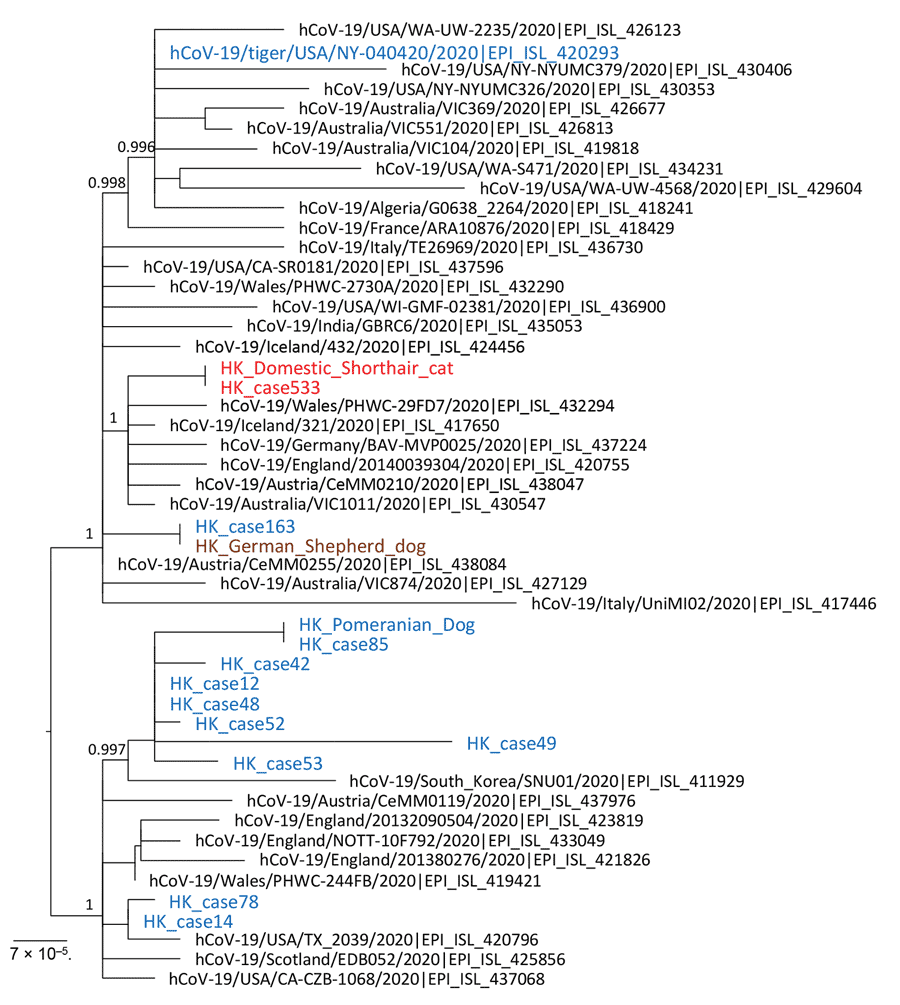
Figure. Phylogenetic analysis of SARS-CoV-2 full genome from an infected cat and the human index case-patient, Hong Kong, China. A virus sequenced directly from a tiger in a zoo in United States was included in this analysis. Virus genome alignment was prepared and manually trimmed at genome 5¢ and 3¢ ends for low-alignment quality. A resulting alignment of 29,655 nt was analyzed by using PhyML (http://www.atgc-montpellier.fr) and the generalized time reversible nucleotide substitution model. Branch support identified by using the fast approximate likelihood ratio test are shown at major nodes. The Hong Kong feline virus from cat 1 and that of its owner are shown in red. Canine and human viruses from Hong Kong, including from the dogs’ owners (HK_case 163 and HK_case 85) are shown in blue. Numbers along branches are bootstrap values. Scale bar indicates nucleotide substitutions per site. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
1These authors contributed equally to this article.