Genomic Epidemiology of Severe Acute Respiratory Syndrome Coronavirus 2, Colombia
Katherine Laiton-Donato

, Christian Julián Villabona-Arenas, José A. Usme-Ciro, Carlos Franco-Muñoz, Diego A. Álvarez-Díaz, Liz Stephany Villabona-Arenas, Susy Echeverría-Londoño, Zulma M. Cucunubá, Nicolás D. Franco-Sierra, Astrid C. Flórez, Carolina Ferro, Nadim J. Ajami, Diana Marcela Walteros, Franklin Prieto, Carlos Andrés Durán, Martha Lucia Ospina-Martínez, and Marcela Mercado-Reyes
Author affiliations: Instituto Nacional de Salud, Bogotá, Colombia (K. Laiton-Donato, J.A. Usme-Ciro, C. Franco-Muñoz, D.A. Álvarez-Díaz, A.C. Flórez, C. Ferro, D.M. Walteros, F. Prieto, C.A. Durán, M.L. Ospina-Martínez, M. Mercado-Reyes); Centre for the Mathematical Modelling of Infectious Diseases (CMMID) and London School of Hygiene & Tropical Medicine, London, UK (C.J. Villabona-Arenas); Universidad Cooperativa de Colombia, Santa Marta, Colombia (J.A. Usme-Ciro); Universidad Industrial de Santander, Bucaramanga, Colombia (L.S. Villabona-Arenas); Imperial College-London, London, UK (S. Echeverría-Londoño, Z.M. Cucunubá); Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Colombia (N. Franco-Sierra); Baylor College of Medicine, Houston, Texas, USA (N.J. Ajami)
Main Article
Figure 3
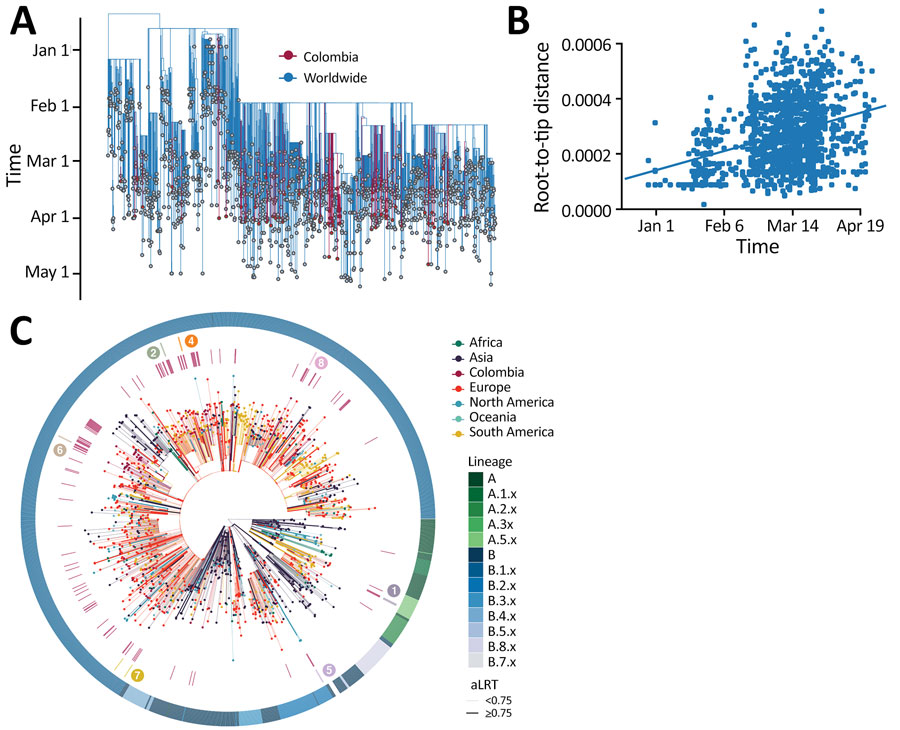
Figure 3. Phylogeny of SARS-CoV-2, Colombia. A) Time-resolved maximum-likelihood tree of 1,578 SARS-CoV-2 sequences. Red indicates 122 sequences from Colombia. B) Root-to-tip distance regression for the sequence data in A. C) Time-resolved maximum-likelihood tree, annotated by region of isolation. The outer ring represents SARS-CoV-2 lineages; the inner red ring highlights the relative position of the sequences from Colombia; the middle ring and the corresponding numbers indicate sequences from epidemiologically linked transmission chains. Branches are colored by the geographic attribution from the migration inference. Highly supported groups are delineated by thicker solid lines. A detailed maximum-likelihood tree is available at https://itol.embl.de/tree/8619015795401231596483440. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Main Article
Page created: September 28, 2020
Page updated: November 23, 2020
Page reviewed: November 23, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
