Volume 26, Number 12—December 2020
Dispatch
Detection and Characterization of Bat Sarbecovirus Phylogenetically Related to SARS-CoV-2, Japan
Figure 1
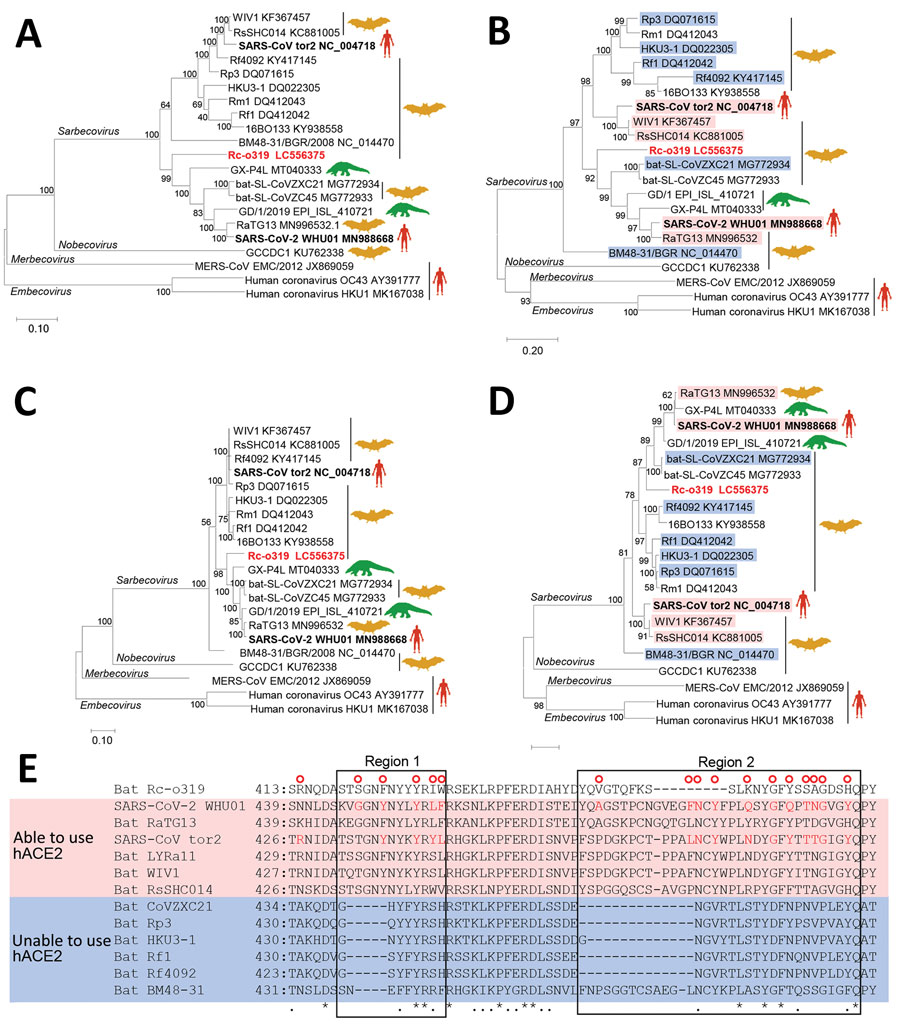
Figure 1. Phylogenetic analysis of sarbecovirus sequenced from little Japanese horseshoe bats (Rhinolophus cornutus) and genetically related to human SARS-CoV-2, Japan. A–D) Phylogenetic trees were generated by using maximum-likelihood analysis combined with 500 bootstrap replicates and show relationships between bat-, human-, and pangolin-derived sarbecoviruses. Phylogenetic trees are shown for nucleotide sequences of the full genome (A), the S protein gene and amino acid sequences (B), the ORF1ab (C), and the S protein (D). Red text indicates positions of Rc-o319, the sarbecovirus sequenced in this study. For panels B and D, magenta bands indicate viruses with S proteins that bind to human ACE2; blue bands indicate viruses with S proteins that do not bind to human ACE2. Bootstrap values are shown above and to the left of the major nodes. Scale bars indicate nucleotide or amino acid substitutions per site. E) Amino acid sequence alignment of the RBM of S proteins that are able or unable to bind to human ACE2. Amino acid residues of the RBM that contact human ACE2 of SARS-CoV-2 and SARS-CoV are indicated in the upper side by red circles. The 2 regions of S protein RBM known to interact with human ACE2 are indicated by boxes labeled region 1 and region 2. ACE2, angiotensin-converting enzyme 2; hACE2, human angiotensin-converting enzyme 2; ORF1ab, open reading frame 1ab; RBM, receptor-binding motif; S, spike protein; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
1These authors contributed equally to this article.