Volume 26, Number 2—February 2020
Research
Novel Subclone of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility, China
Figure 2
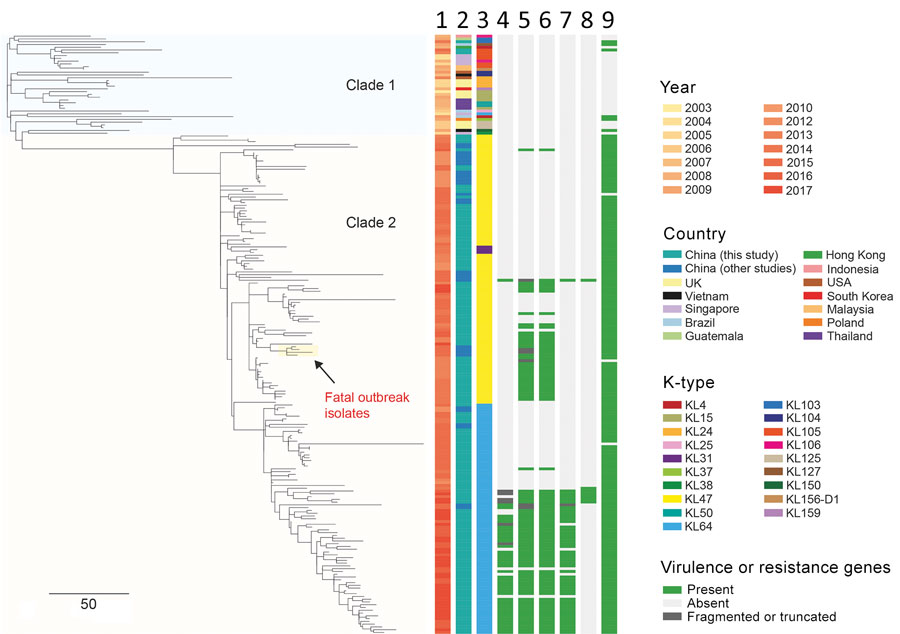
Figure 2. Phylogenetic analysis of 216 CRKP ST11 isolates, China, 2013–2017, including 154 CRKP isolates collected during 2012–2017 in study of bloodstream infections in a tertiary hospital and 62 isolates that were sequenced in previous studies (Appendix 2 Table 1). The phylogenetic tree was obtained by mapping all sequence reads to the hybrid assembly of KP47434 and removing the recombined regions from the alignment. The tree was rooted using ST1731 isolate EuSCAPE_ES29 (ERR1541319), which was included in this analysis but later removed from the tree (a tree including this outgroup is shown in Appendix 1 Figure 1). Five capsular types (KL31, KL47, KL64, KL103, and KL105) were detected in our ST11 collection, which are indicated in different colors as shown in the legend. Some of virulence genes detected are shown here. The rmpA2 gene carried by KL64 isolates was frameshifted, namely rmpA2*. Aerobactin and salmochelin represent the iucABCD-iutA and iroBCDN gene clusters, respectively. The fatal outbreak clone reported in China recently (12) is highlighted on the tree. Lanes: 1, year; 2, country; 3, K-type; 4, rmpA; 5, rmpA2; 6, aerobactin; 7, peg-344; 8, salmochelin; 9, blaKPC. Scale bar indicates single-nucleotide polymorphisms. CRKP, carbapenem-resistant Klebsiella pneumoniae; KL, capsular loci; ST, sequence type.
References
- Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA. 2015;314:1479–87. DOIPubMedGoogle Scholar
- Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasević AT, et al.; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17:153–63. DOIPubMedGoogle Scholar
- Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. DOIPubMedGoogle Scholar
- Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–106. DOIPubMedGoogle Scholar
- Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–84. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Pitout JDD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 2014;5:e01355–14. DOIPubMedGoogle Scholar
- Gaiarsa S, Comandatore F, Gaibani P, Corbella M, Dalla Valle C, Epis S, et al. Genomic epidemiology of Klebsiella pneumoniae in Italy and novel insights into the origin and global evolution of its resistance to carbapenem antibiotics. Antimicrob Agents Chemother. 2015;59:389–96. DOIPubMedGoogle Scholar
- Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014;111:4988–93. DOIPubMedGoogle Scholar
- Wyres KL, Gorrie C, Edwards DJ, Wertheim HFL, Hsu LY, Van Kinh N, et al. Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol Evol. 2015;7:1267–79. DOIPubMedGoogle Scholar
- Jiang Y, Wei Z, Wang Y, Hua X, Feng Y, Yu Y. Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole genome sequencing. Clin Microbiol Infect. 2015;21:1001–7. DOIPubMedGoogle Scholar
- Dong N, Zhang R, Liu L, Li R, Lin D, Chan EW-C, et al. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microb Genom. 2018;4:54–12. DOIPubMedGoogle Scholar
- Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. DOIPubMedGoogle Scholar
- Du P, Zhang Y, Chen C. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18:23–4. DOIPubMedGoogle Scholar
- Yao H, Qin S, Chen S, Shen J, Du X-D. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18:25. DOIPubMedGoogle Scholar
- Wong MHY, Shum H-P, Chen JHK, Man M-Y, Wu A, Chan EW-C, et al. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18:24. DOIPubMedGoogle Scholar
- Xiao T, Yu W, Niu T, Huang C, Xiao Y. A retrospective, comparative analysis of risk factors and outcomes in carbapenem-susceptible and carbapenem-nonsusceptible Klebsiella pneumoniae bloodstream infections: tigecycline significantly increases the mortality. Infect Drug Resist. 2018;11:595–606. DOIPubMedGoogle Scholar
- Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–82. DOIPubMedGoogle Scholar
- McLaughlin MM, Advincula MR, Malczynski M, Barajas G, Qi C, Scheetz MH. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a Galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect Dis. 2014;14:31. DOIPubMedGoogle Scholar
- Naparstek L, Carmeli Y, Navon-Venezia S, Banin E. Biofilm formation and susceptibility to gentamicin and colistin of extremely drug-resistant KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2014;69:1027–34. DOIPubMedGoogle Scholar
- Wang L, Shen D, Wu H, Ma Y. Resistance of hypervirulent Klebsiella pneumoniae to both intracellular and extracellular killing of neutrophils. PLoS One. 2017;12:
e0173638 . DOIPubMedGoogle Scholar - Havill NL, Boyce JM, Otter JA. Extended survival of carbapenem-resistant Enterobacteriaceae on dry surfaces. Infect Control Hosp Epidemiol. 2014;35:445–7. DOIPubMedGoogle Scholar
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Henson SP, Boinett CJ, Ellington MJ, Kagia N, Mwarumba S, Nyongesa S, et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017;307:422–9. DOIPubMedGoogle Scholar
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. DOIPubMedGoogle Scholar
- Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One. 2015;10:
e0133727 . DOIPubMedGoogle Scholar - Moradigaravand D, Martin V, Peacock SJ, Parkhill J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio. 2017;8:e01976–16. DOIPubMedGoogle Scholar
- Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46:e134–4. DOIPubMedGoogle Scholar
- Lee E-M, Ahn S-H, Park J-H, Lee J-H, Ahn S-C, Kong I-S. Identification of oligopeptide permease (opp) gene cluster in Vibrio fluvialis and characterization of biofilm production by oppA knockout mutation. FEMS Microbiol Lett. 2004;240:21–30. DOIPubMedGoogle Scholar
- Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, Jarillo-Quijada MD, von Bargen K, Torres J, et al. H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front Cell Infect Microbiol. 2016;6:13. DOIPubMedGoogle Scholar
- Kobayashi SD, Porter AR, Freedman B, Pandey R, Chen L, Kreiswirth BN, et al. Antibody-mediated killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. MBio. 2018;9:1198. DOIPubMedGoogle Scholar
- Pan Y-J, Lin T-L, Lin Y-T, Su P-A, Chen C-T, Hsieh P-F, et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother. 2015;59:1038–47. DOIPubMedGoogle Scholar
- Koh TH, Cao D, Shan QY, Bacon A, Hsu LY, Ooi EE. Acquired carbapenemases in Enterobactericeae in Singapore, 1996-2012. Pathology. 2013;45:600–3. DOIPubMedGoogle Scholar
- Cryz SJ Jr, Mortimer PM, Mansfield V, Germanier R. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J Clin Microbiol. 1986;23:687–90. DOIPubMedGoogle Scholar
- Jones CL, Clancy M, Honnold C, Singh S, Snesrud E, Onmus-Leone F, et al. Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin Infect Dis. 2015;61:145–54. DOIPubMedGoogle Scholar
- Melin M, Trzciński K, Meri S, Käyhty H, Väkeväinen M. The capsular serotype of Streptococcus pneumoniae is more important than the genetic background for resistance to complement. Infect Immun. 2010;78:5262–70. DOIPubMedGoogle Scholar
- Rukke HV, Kalluru RS, Repnik U, Gerlini A, José RJ, Periselneris J, et al. Protective role of the capsule and impact of serotype 4 switching on Streptococcus mitis. Infect Immun. 2014;82:3790–801. DOIPubMedGoogle Scholar
- Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio. 2015;6:
e00630 . DOIPubMedGoogle Scholar - Hsu C-R, Lin T-L, Chen Y-C, Chou H-C, Wang J-T. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157:3446–57. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.