Volume 26, Number 6—June 2020
Dispatch
Prevalence of Escherichia albertii in Raccoons (Procyon lotor), Japan
Figure
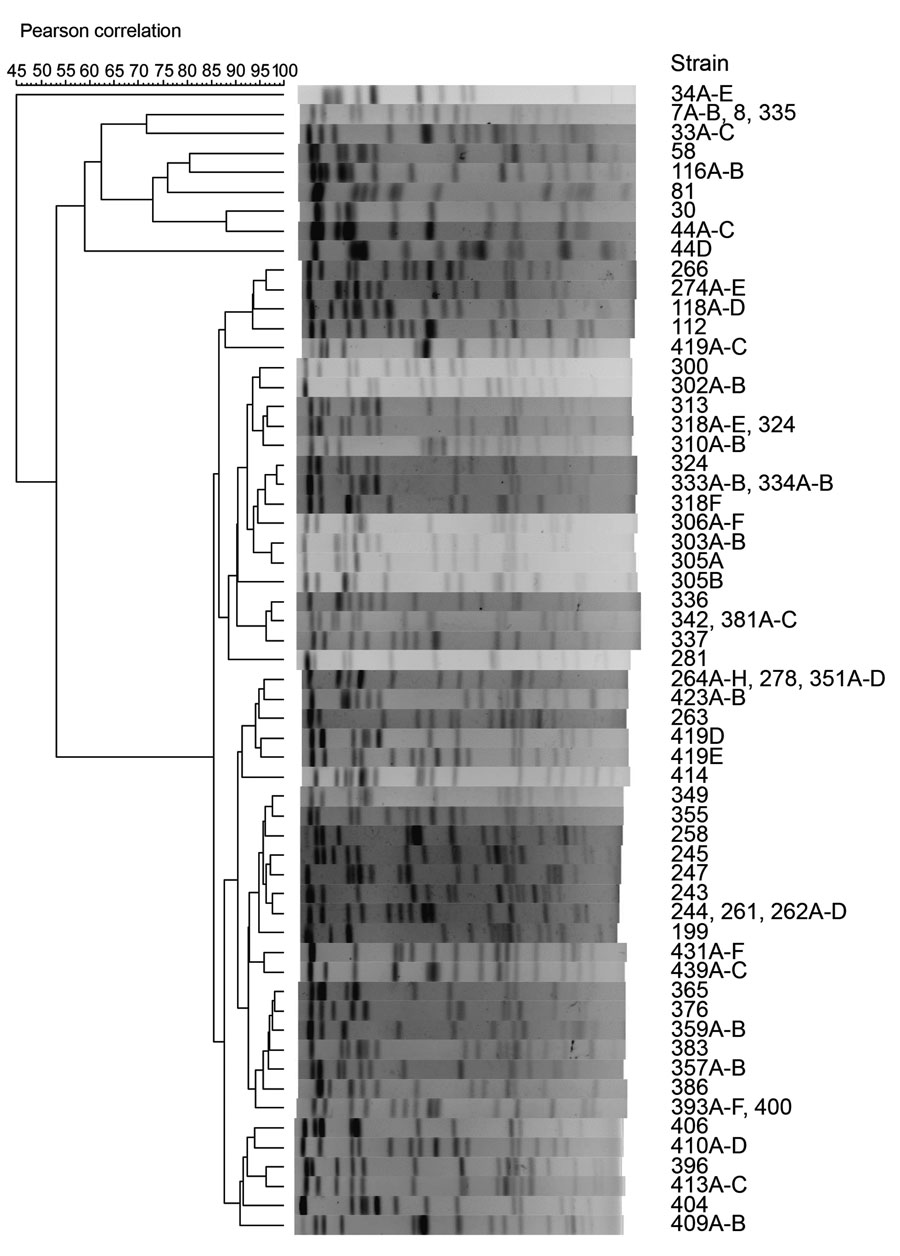
Figure. Phylogenetic analysis of raccoon Escherichia albertii strains by pulsed-field gel electrophoresis (PFGE). XbaI-digested genomic DNA of 143 raccoon E. albertii strains isolated in this study were analyzed by PFGE. The dendrogram was constructed based on DNA fingerprints obtained (Appendix). The number in each strain name represents a specific raccoon identification number.
Page created: May 19, 2020
Page updated: May 19, 2020
Page reviewed: May 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.