Volume 26, Number 7—July 2020
Research
Efficient Surveillance of Plasmodium knowlesi Genetic Subpopulations, Malaysian Borneo, 2000–2018
Figure 2
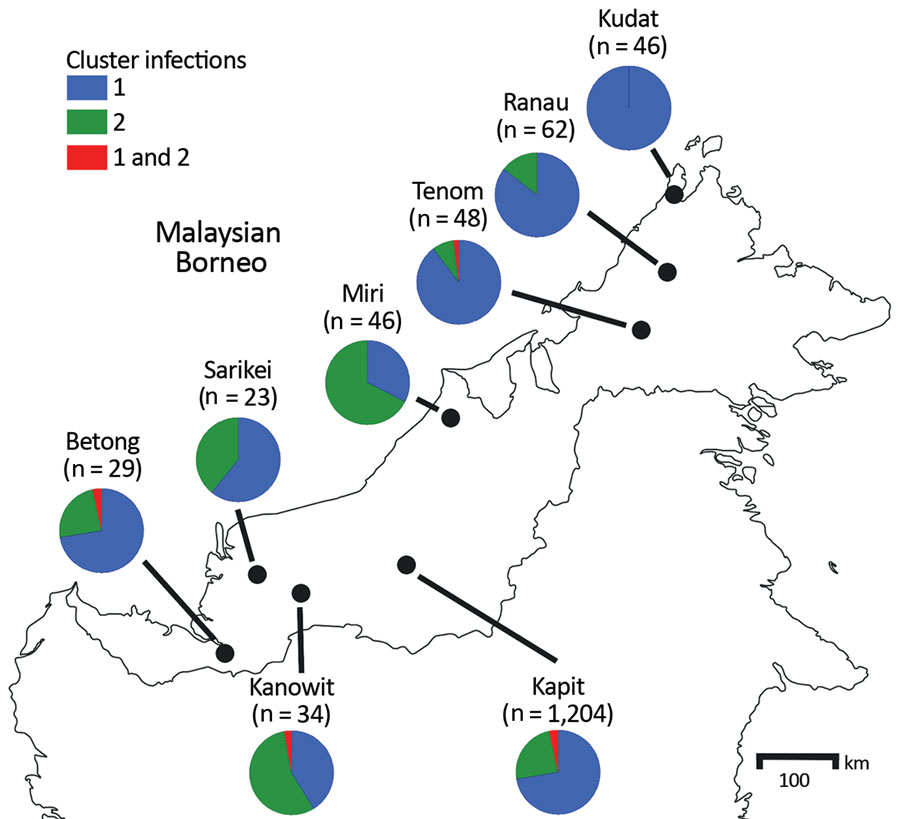
Figure 2. Proportions of cluster 1 and cluster 2 Plasmodium knowlesi infections as defined with the cluster-specific PCRs, Malaysian Borneo. A total of 1,492 P. knowlesi infections from humans were genotyped using the cluster-specific PCR primers. The exact numbers for each location are shown in Appendix Table 3.
1These authors contributed equally to this article.
Page created: April 10, 2020
Page updated: June 18, 2020
Page reviewed: June 18, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.