Volume 26, Number 7—July 2020
Dispatch
Carbapenem Resistance Conferred by OXA-48 in K2-ST86 Hypervirulent Klebsiella pneumoniae, France
Figure 1
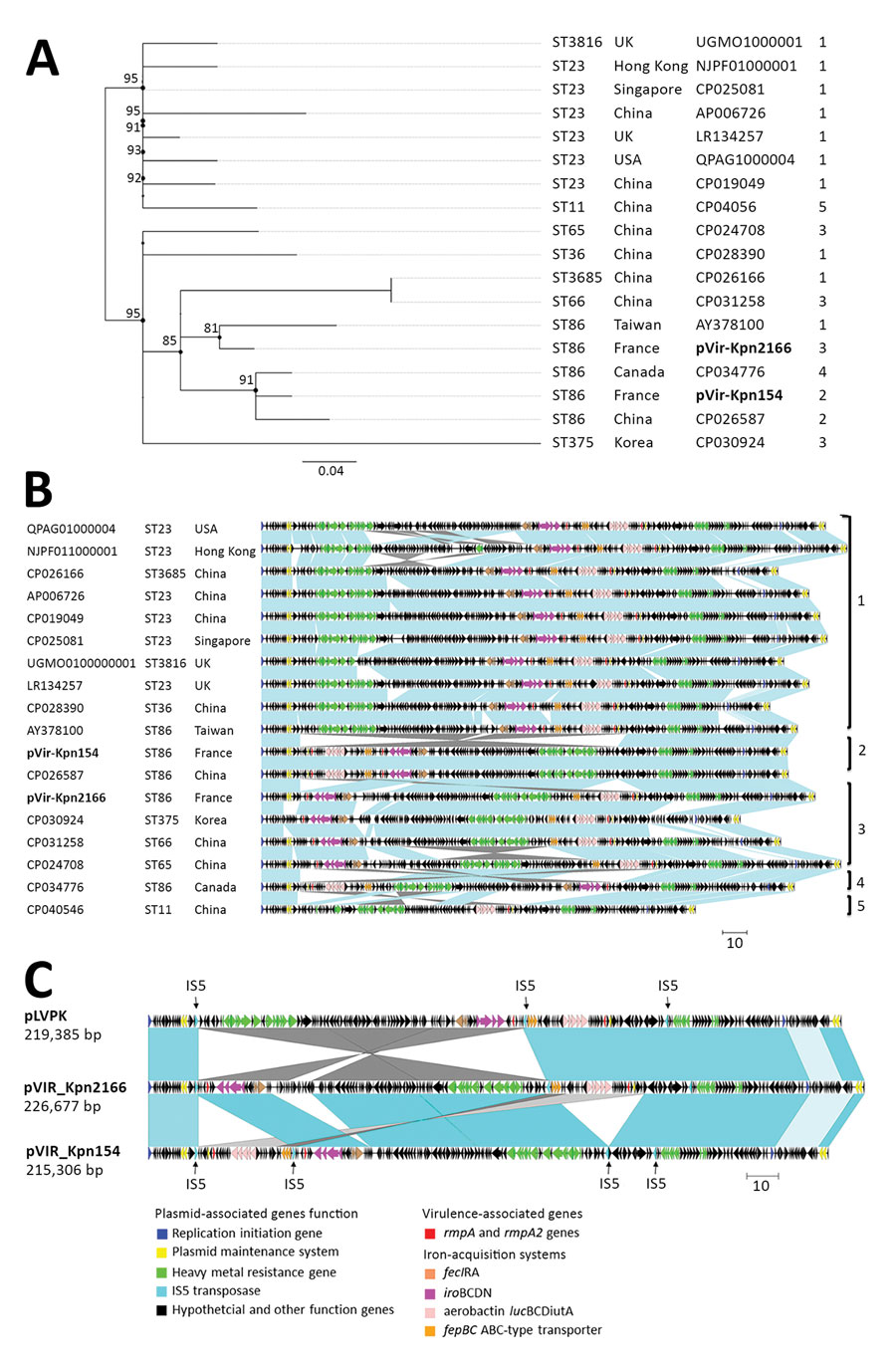
Figure 1. Comparison of pVIR-Kpn2166 and pVIR-Kpn154 Klebsiella pneumoniae isolates from 2 patients in France (bold) with 16 hypervirulent K. pneumoniae virulence plasmids recovered from the PATRIC database (http://www.patricbrc.org). A) Single-nucleotide polymorphism–based phylogenetic tree built by RaxML from an alignment generated by Burrows-Wheeler Aligner and filtered to remove recombination using Gubbins as previously described (9). The ST and the geographic origin of bacterial hosts are shown. Scale bar indicates mean number of nucleotide substitutions per site. B) Synteny analysis of hypervirulent K. pneumoniae virulence plasmids based on data from blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Virulence-based synteny groups are indicated and the operons encoding the virulence factors, virulence synteny groups of plasmids and the ST and the geographic origin of bacterial hosts. Scale bar indicates kbp. C) Comparison of the details of rearrangements observed in pVIR-Kpn2166 and pVIR-Kpn154 and in pLVPK. Scale bar indicates kbp. ST, sequence type.
References
- Meatherall BL, Gregson D, Ross T, Pitout JDD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med. 2009;122:866–73. DOIPubMedGoogle Scholar
- Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:
e1008114 . DOIPubMedGoogle Scholar - Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EME, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2016;16:e190–5. DOIPubMedGoogle Scholar
- Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–18. DOIPubMedGoogle Scholar
- Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4. DOIPubMedGoogle Scholar
- Villa L, Feudi C, Fortini D, García-Fernández A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother. 2014;58:1707–12. DOIPubMedGoogle Scholar
- Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–98. DOIPubMedGoogle Scholar
- Beyrouthy R, Barets M, Marion E, Dananché C, Dauwalder O, Robin F, et al. Novel enterobacter lineage as leading cause of nosocomial outbreak involving carbapenemase-producing strains. Emerg Infect Dis. 2018;24:1505–15. DOIPubMedGoogle Scholar
- Beyrouthy R, Robin F, Cougnoux A, Dalmasso G, Darfeuille-Michaud A, Mallat H, et al. Chromosome-mediated OXA-48 carbapenemase in highly virulent Escherichia coli. J Antimicrob Chemother. 2013;68:1558–61. DOIPubMedGoogle Scholar
- Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56:1–12. DOIPubMedGoogle Scholar
- Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157:3446–57. DOIPubMedGoogle Scholar
- Mataseje LF, Boyd DA, Mulvey MR, Longtin Y. Two hypervirulent Klebsiella pneumoniae isolates producing a blaKPC-2 carbapenemase from a Canadian patient. Antimicrob Agents Chemother. 2019;63:1–4. DOIPubMedGoogle Scholar
- Karlsson M, Stanton RA, Ansari U, McAllister G, Chan MY, Sula E, et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob Agents Chemother. 2019;63:1–6. DOIPubMedGoogle Scholar
- Roulston KJ, Bharucha T, Turton JF, Hopkins KL, Mack DJF. A case of NDM-carbapenemase-producing hypervirulent Klebsiella pneumoniae sequence type 23 from the UK. JMM Case Rep. 2018;5:
e005130 . DOIPubMedGoogle Scholar