Hepatitis E Virus Genotype 7 RNA and Antibody Kinetics in Naturally Infected Dromedary Calves, United Arab Emirates
Victor M. Corman

, Peter Nagy, Stefanie Ostermann, Jacqueline Arloth, Anne Liljander, Rajib Barua, Aungshuman Das Gupta, Fatima Hakimuddin, Judit Juhasz, Ulrich Wernery, and Christian Drosten
Author affiliations: Charité-Universitätsmedizin Berlin, Berlin, Germany. (V.M. Corman, C. Drosten); German Centre for Infection Research, Berlin (V.M. Corman, C. Drosten); Emirates Industries for Camel Milk and Products, Dubai, United Arab Emirates (P. Nagy, R. Barua, A. Das Gupta, J. Juhasz); Institute of Experimental Immunology, Lübeck, Germany (S. Ostermann, J. Arloth, A. Liljander); Central Veterinary Research Laboratory, Dubai (F. Hakimuddin, U. Wernery)
Main Article
Figure 2
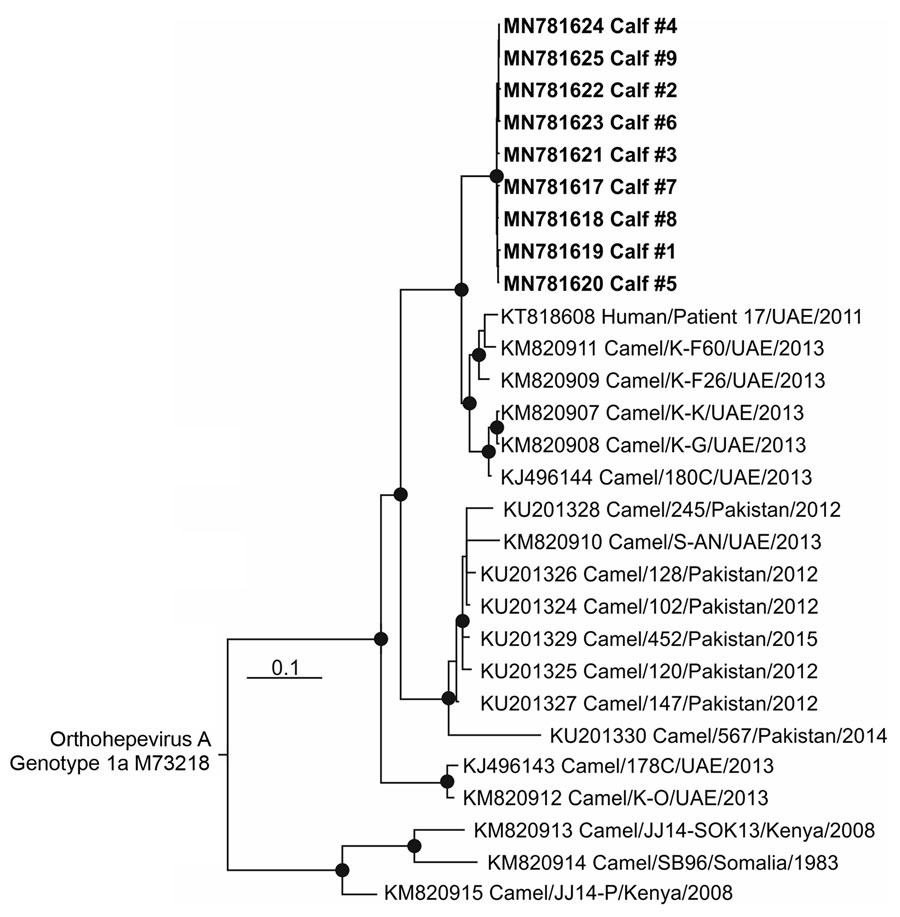
Figure 2. Phylogenetic tree of open reading frame 1 of HEV RNA sequences from 11 camel calves (bold), United Arab Emirates, and other Orthohepevirus A genotype 7 sequences available in GenBank as of May 1, 2019. The tree was calculated by using MrBayes (http://mrbayes.sourceforge.net) and a generalized time reversible substitution model. One million generations were sampled every 500 steps, and 20% of trees were discarded as burn-in. Bold indicates sequences obtained during this study. An Orthohepevirus A genotype 1 sequence (GenBank accession no M73218) was used as the outgroup. Taxon names of all reference sequences include GenBank accession number, strain, country of origin, and year of sampling. GenBank accession numbers for sequences obtained in this study are MN781617–25. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: May 04, 2020
Page updated: August 19, 2020
Page reviewed: August 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
